Geographic structuring of the Plasmodium falciparum sarco(endo)plasmic reticulum Ca2+ ATPase (PfSERCA) gene diversity
- PMID: 20195531
- PMCID: PMC2828472
- DOI: 10.1371/journal.pone.0009424
Geographic structuring of the Plasmodium falciparum sarco(endo)plasmic reticulum Ca2+ ATPase (PfSERCA) gene diversity
Abstract
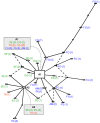
Artemisinin, a thapsigargin-like sesquiterpene has been shown to inhibit the Plasmodium falciparum sarco/endoplasmic reticulum calcium-ATPase PfSERCA. To collect baseline pfserca sequence information before field deployment of Artemisinin-based Combination therapies that may select mutant parasites, we conducted a sequence analysis of 100 isolates from multiple sites in Africa, Asia and South America. Coding sequence diversity was large, with 29 mutated codons, including 32 SNPs (average of one SNP/115 bp), of which 19 were novel mutations. Most SNP detected in this study were clustered within a region in the cytosolic head of the protein. The PfSERCA functional domains were very well conserved, with non synonymous mutations located outside the functional domains, except for the S769N mutation associated in French Guiana with elevated IC(50) for artemether. The S769N mutation is located close to the hinge of the headpiece, which in other species modulates calcium affinity and in consequence efficacy of inhibitors, possibly linking calcium homeostasis to drug resistance. Genetic diversity was highest in Senegal, Brazil and French Guiana, and few mutations were identified in Asia. Population genetic analysis was conducted for a partial fragment of the gene encompassing nucleotide coordinates 87-2862 (unambiguous sequence available for 96 isolates). This supported a geographic clustering, with a separation between Old and New World samples and one dominant ancestral haplotype. Genetic drift alone cannot explain the observed polymorphism, suggesting that other evolutionary mechanisms are operating. One possible contributor could be the frequency of haemoglobinopathies that are associated with calcium dysregulation in the erythrocyte.
Conflict of interest statement
Figures

References
-
- World Malaria Report. (2008) WHO. www.who.int.
-
- Jambou R, Legrand E, Niang M, Khim N, Lim P, et al. Resistance of Plasmodium falciparum field isolates to in-vitro artemether and point mutations of the SERCA-type PfATPase6. Lancet. 2005;366:1960–1963. - PubMed
-
- Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM. Artemisinin Resistance in Cambodia 1 (ARC1) Study Consortium Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359:2619–20. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

