The genome of Streptococcus mitis B6--what is a commensal?
- PMID: 20195536
- PMCID: PMC2828477
- DOI: 10.1371/journal.pone.0009426
The genome of Streptococcus mitis B6--what is a commensal?
Abstract
Streptococcus mitis is the closest relative of the major human pathogen S. pneumoniae. The 2,15 Mb sequence of the Streptococcus mitis B6 chromosome, an unusually high-level beta-lactam resistant and multiple antibiotic resistant strain, has now been determined to encode 2100 genes. The accessory genome is estimated to represent over 40%, including 75 mostly novel transposases and IS, the prophage phiB6 and another seven phage related regions. Tetracycline resistance mediated by Tn5801, and an unusual and large gene cluster containing three aminoglycoside resistance determinants have not been described in other Streptococcus spp. Comparative genomic analyses including hybridization experiments on a S. mitis B6 specific microarray reveal that individual S. mitis strains are almost as distantly related to the B6 strain as S. pneumoniae. Both species share a core of over 900 genes. Most proteins described as pneumococcal virulence factors are present in S. mitis B6, but the three choline binding proteins PcpA, PspA and PspC, and three gene clusters containing the hyaluronidase gene, ply and lytA, and the capsular genes are absent in S. mitis B6 and other S. mitis as well and confirm their importance for the pathogenetic potential of S. pneumoniae. Despite the close relatedness between the two species, the S. mitis B6 genome reveals a striking X-alignment when compared with S. pneumoniae.
Conflict of interest statement
Figures
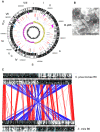
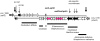
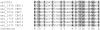

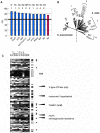
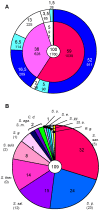

Similar articles
-
Discrimination of Streptococcus pneumoniae from viridans group streptococci by genomic subtractive hybridization.J Clin Microbiol. 2005 Sep;43(9):4528-34. doi: 10.1128/JCM.43.9.4528-4534.2005. J Clin Microbiol. 2005. PMID: 16145102 Free PMC article.
-
Identification of Virulence-Associated Properties by Comparative Genome Analysis of Streptococcus pneumoniae, S. pseudopneumoniae, S. mitis, Three S. oralis Subspecies, and S. infantis.mBio. 2019 Sep 3;10(5):e01985-19. doi: 10.1128/mBio.01985-19. mBio. 2019. PMID: 31481387 Free PMC article.
-
Versatility of choline metabolism and choline-binding proteins in Streptococcus pneumoniae and commensal streptococci.FEMS Microbiol Rev. 2009 May;33(3):572-86. doi: 10.1111/j.1574-6976.2009.00172.x. FEMS Microbiol Rev. 2009. PMID: 19396958 Review.
-
Identification of a secreted cholesterol-dependent cytolysin (mitilysin) from Streptococcus mitis.J Bacteriol. 2007 Jan;189(2):627-32. doi: 10.1128/JB.01092-06. Epub 2006 Oct 27. J Bacteriol. 2007. PMID: 17071760 Free PMC article.
-
Streptococcus mitis: walking the line between commensalism and pathogenesis.Mol Oral Microbiol. 2011 Apr;26(2):89-98. doi: 10.1111/j.2041-1014.2010.00601.x. Epub 2011 Jan 18. Mol Oral Microbiol. 2011. PMID: 21375700 Review.
Cited by
-
A high-resolution view of genome-wide pneumococcal transformation.PLoS Pathog. 2012;8(6):e1002745. doi: 10.1371/journal.ppat.1002745. Epub 2012 Jun 14. PLoS Pathog. 2012. PMID: 22719250 Free PMC article.
-
Identification, variation and transcription of pneumococcal repeat sequences.BMC Genomics. 2011 Feb 18;12:120. doi: 10.1186/1471-2164-12-120. BMC Genomics. 2011. PMID: 21333003 Free PMC article.
-
Comparison of alternative mixture model methods to analyze bacterial CGH experiments with multi-genome arrays.BMC Res Notes. 2014 Mar 14;7:148. doi: 10.1186/1756-0500-7-148. BMC Res Notes. 2014. PMID: 24629208 Free PMC article.
-
Large scale genomic analysis shows no evidence for pathogen adaptation between the blood and cerebrospinal fluid niches during bacterial meningitis.Microb Genom. 2017 Jan 31;3(1):e000103. doi: 10.1099/mgen.0.000103. eCollection 2017 Jan. Microb Genom. 2017. PMID: 28348877 Free PMC article.
-
Evidence of antimicrobial resistance-conferring genetic elements among pneumococci isolated prior to 1974.BMC Genomics. 2013 Jul 24;14:500. doi: 10.1186/1471-2164-14-500. BMC Genomics. 2013. PMID: 23879707 Free PMC article.
References
-
- Bourgault AM, Wilson WR, Washington JA II. Antimicrobial susceptibilities of species of viridans streptococci. J Infect Dis. 1979;140:316–324. - PubMed
-
- Brandenburg RO, Giuliani ER, Wilson WR, Geraci JE. Infective endocarditis - a 25-year overview of diagnosis and therapy. J Am Coll Cardiol. 1983;1:280–291. - PubMed
-
- Van der Meer JTM, van Vianen W, Hu E, van Leeuwen WB, Valkenburg HA, et al. Distribution, antibiotic susceptibility and tolerance of bacterial isolates in culture-positive cases of endocarditis in The Netherlands. Eur J Clin Microbiol Infect Dis. 1991;10:728–734. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

