A flexible docking scheme to explore the binding selectivity of PDZ domains
- PMID: 20196074
- PMCID: PMC2868235
- DOI: 10.1002/pro.366
A flexible docking scheme to explore the binding selectivity of PDZ domains
Abstract
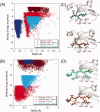
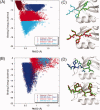
Modeling of protein binding site flexibility in molecular docking is still a challenging problem due to the large conformational space that needs sampling. Here, we propose a flexible receptor docking scheme: A dihedral restrained replica exchange molecular dynamics (REMD), where we incorporate the normal modes obtained by the Elastic Network Model (ENM) as dihedral restraints to speed up the search towards correct binding site conformations. To our knowledge, this is the first approach that uses ENM modes to bias REMD simulations towards binding induced fluctuations in docking studies. In our docking scheme, we first obtain the deformed structures of the unbound protein as initial conformations by moving along the binding fluctuation mode, and perform REMD using the ENM modes as dihedral restraints. Then, we generate an ensemble of multiple receptor conformations (MRCs) by clustering the lowest replica trajectory. Using ROSETTALIGAND, we dock ligands to the clustered conformations to predict the binding pose and affinity. We apply this method to postsynaptic density-95/Dlg/ZO-1 (PDZ) domains; whose dynamics govern their binding specificity. Our approach produces the lowest energy bound complexes with an average ligand root mean square deviation of 0.36 A. We further test our method on (i) homologs and (ii) mutant structures of PDZ where mutations alter the binding selectivity. In both cases, our approach succeeds to predict the correct pose and the affinity of binding peptides. Overall, with this approach, we generate an ensemble of MRCs that leads to predict the binding poses and specificities of a protein complex accurately.
Figures

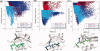
References
-
- Halperin I, Ma B, Wolfson H, Nussinov R. Principles of docking: an overview of search algorithms and a guide to scoring functions. Proteins: Struct Funct Bioinf. 2002;47:409–443. - PubMed
-
- Leach AR. Molecular Modelling: Principles and Applications. New York: Prentice Hall; 2001.
-
- Mohan V, Gibbs AC, Cummings MD, Jaeger EP, DesJarlais RL. Docking: successes and challenges. Curr Pharm Des. 2005;11:323–333. - PubMed
-
- Sousa SF, Fernandes PA, Ramos MJ. Protein-ligand docking: current status and future challenges. Proteins: Struct Funct Bioinf. 2006;65:15–26. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

