Localization of MglA, an essential gliding motility protein in Myxococcus xanthus
- PMID: 20196075
- PMCID: PMC3225289
- DOI: 10.1002/cm.20447
Localization of MglA, an essential gliding motility protein in Myxococcus xanthus
Abstract
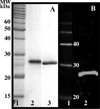
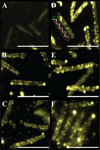
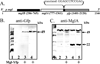
MglA, a 22-kDa protein related to monomeric GTPases, is required for the normal operation of the A (Adventurous) and S (Social) motility and for multicellular development of Myxococcus xanthus. To determine how MglA controls A- and S-motility, MglA was assayed biochemically and its cellular location was determined. His-tagged MglA hydrolyzed GTP slowly in vitro at a rate nearly identical to that of Ras showing that MglA has GTPase activity. Immunofluorescence microscopy of fixed cells from liquid showed that MglA was associated with helical track similar to the MreB spiral that spanned the length of the cell. The distribution pattern of MglA depended on the type of surface from which cells were harvested. In cells gliding on 1.5% (w/v) agar, the helical pattern gave way to punctate clusters of MglA-Yfp at the poles and along the long axis (lateral clusters). The lateral clusters emerged near the leading pole as the cell advanced coincident with a decrease in the intensity of the MglA-Yfp cluster at the leading pole. Newly formed lateral clusters remained fixed with regard to the substratum as the cell moved forward, similar to focal adhesion complexes described for AglZ, a protein partner of MglA. Lateral clusters did not form in cells gliding in methylcellulose, a polymer that stimulates S-motility at low cell density; rather MglA-Yfp was diffuse in the cytoplasm and more concentrated at the poles. The results suggest that conditions that favor S-motility prevent the formation of lateral clusters of MglA, which are associated with A-motility functions.
2010 Wiley-Liss, Inc.
Figures





Similar articles
-
Bacterial motility complexes require the actin-like protein, MreB and the Ras homologue, MglA.EMBO J. 2010 Jan 20;29(2):315-26. doi: 10.1038/emboj.2009.356. Epub 2009 Dec 3. EMBO J. 2010. PMID: 19959988 Free PMC article.
-
A response regulator interfaces between the Frz chemosensory system and the MglA/MglB GTPase/GAP module to regulate polarity in Myxococcus xanthus.PLoS Genet. 2012 Sep;8(9):e1002951. doi: 10.1371/journal.pgen.1002951. Epub 2012 Sep 13. PLoS Genet. 2012. PMID: 23028358 Free PMC article.
-
The small G-protein MglA connects to the MreB actin cytoskeleton at bacterial focal adhesions.J Cell Biol. 2015 Jul 20;210(2):243-56. doi: 10.1083/jcb.201412047. Epub 2015 Jul 13. J Cell Biol. 2015. PMID: 26169353 Free PMC article.
-
Gliding motility in bacteria: insights from studies of Myxococcus xanthus.Microbiol Mol Biol Rev. 1999 Sep;63(3):621-41. doi: 10.1128/MMBR.63.3.621-641.1999. Microbiol Mol Biol Rev. 1999. PMID: 10477310 Free PMC article. Review.
-
The mysterious nature of bacterial surface (gliding) motility: A focal adhesion-based mechanism in Myxococcus xanthus.Semin Cell Dev Biol. 2015 Oct;46:143-54. doi: 10.1016/j.semcdb.2015.10.033. Epub 2015 Oct 28. Semin Cell Dev Biol. 2015. PMID: 26520023 Review.
Cited by
-
Cell polarity/motility in bacteria: closer to eukaryotes than expected?EMBO J. 2010 Jul 21;29(14):2258-9. doi: 10.1038/emboj.2010.144. EMBO J. 2010. PMID: 20648047 Free PMC article. No abstract available.
-
Structural analysis of the Ras-like G protein MglA and its cognate GAP MglB and implications for bacterial polarity.EMBO J. 2011 Aug 16;30(20):4185-97. doi: 10.1038/emboj.2011.291. EMBO J. 2011. PMID: 21847100 Free PMC article.
-
Global analysis of phase variation in Myxococcus xanthus.Mol Microbiol. 2011 Aug;81(3):784-804. doi: 10.1111/j.1365-2958.2011.07732.x. Epub 2011 Jul 4. Mol Microbiol. 2011. PMID: 21722202 Free PMC article.
-
Effects of site-directed mutagenesis of mglA on motility and swarming of Myxococcus xanthus.BMC Microbiol. 2010 Nov 18;10:295. doi: 10.1186/1471-2180-10-295. BMC Microbiol. 2010. PMID: 21083931 Free PMC article.
-
Mathematical modeling of mechanosensitive reversal control in Myxococcus xanthus.Front Microbiol. 2024 Jan 8;14:1294631. doi: 10.3389/fmicb.2023.1294631. eCollection 2023. Front Microbiol. 2024. PMID: 38260904 Free PMC article.
References
-
- Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science. 2008;320(5883):1636–1638. - PubMed
-
- Bowden MG, Kaplan HB. The Myxococcus xanthus lipopolysaccharide O-antigen is required for social motility and multicellular development. Mol. Microbiol. 1998;30(2):275–284. - PubMed
-
- Chen JC, Beckwith J. FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division. Mol Microbiol. 2001;42(2):395–413. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

