Progress in phage display: evolution of the technique and its application
- PMID: 20196239
- PMCID: PMC11115567
- DOI: 10.1007/s00018-009-0192-2
Progress in phage display: evolution of the technique and its application
Abstract
Phage display, the presentation of (poly)peptides as fusions to capsid proteins on the surface of bacterial viruses, celebrates its 25th birthday in 2010. The technique, coupled with in vitro selection, enables rapid identification and optimization of proteins based on their structural or functional properties. In the last two decades, it has advanced tremendously and has become widely accepted by the scientific community. This by no means exhaustive review aims to inform the reader of the key modifications in phage display. Novel display formats, innovative library designs and screening strategies are discussed. I will also briefly review some recent uses of the technology to illustrate its incredible versatility.
Figures
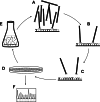

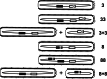
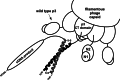
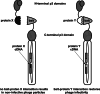
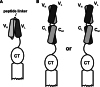
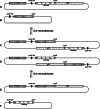
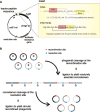
References
-
- Pennazio S. The origin of phage virology. Riv Biol. 2006;99:103–129. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

