Modular structure of smooth muscle Myosin light chain kinase: hydrodynamic modeling and functional implications
- PMID: 20196616
- PMCID: PMC2864612
- DOI: 10.1021/bi901963e
Modular structure of smooth muscle Myosin light chain kinase: hydrodynamic modeling and functional implications
Abstract
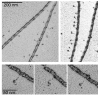
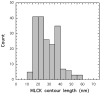
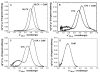
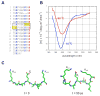
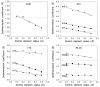
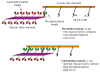
Smooth muscle myosin light chain kinase (smMLCK) is a calcium-calmodulin complex-dependent enzyme that activates contraction of smooth muscle. The polypeptide chain of rabbit uterine smMLCK (Swiss-Prot entry P29294) contains the catalytic/regulatory domain, three immunoglobulin-related motifs (Ig), one fibronectin-related motif (Fn3), a repetitive, proline-rich segment (PEVK), and, at the N-terminus, a unique F-actin-binding domain. We have evaluated the spatial arrangement of these domains in a recombinant 125 kDa full-length smMLCK and its two catalytically active C-terminal fragments (77 kDa, residues 461-1147, and 61 kDa, residues 461-1002). Electron microscopic images of smMLCK cross-linked to F-actin show particles at variable distances (11-55 nm) from the filament, suggesting that a well-structured C-terminal segment of smMLCK is connected to the actin-binding domain by a long, flexible tether. We have used structural homology and molecular dynamics methods to construct various all-atom representation models of smMLCK and its two fragments. The theoretical sedimentation coefficients computed with HYDROPRO were compared with those determined by sedimentation velocity. We found agreement between the predicted and observed sedimentation coefficients for models in which the independently folded catalytic domain, Fn3, and Ig domains are aligned consecutively on the long axis of the molecule. The PEVK segment is modeled as an extensible linker that enables smMLCK to remain bound to F-actin and simultaneously activate the myosin heads of adjacent myosin filaments at a distance of >or=40 nm. The structural properties of smMLCK may contribute to the elasticity of smooth muscle cells.
Figures






Similar articles
-
Calmodulin kinase II chimeras used to investigate the structural requirements for smooth muscle myosin light chain kinase autoinhibition and calmodulin-dependent activation.Biochemistry. 1999 Nov 16;38(46):15061-9. doi: 10.1021/bi990883a. Biochemistry. 1999. PMID: 10563788
-
Myosin light chain kinase: functional domains and structural motifs.Acta Physiol Scand. 1998 Dec;164(4):471-82. doi: 10.1111/j.1365-201x.1998.tb10699.x. Acta Physiol Scand. 1998. PMID: 9887970 Review.
-
Identification of a novel actin binding motif in smooth muscle myosin light chain kinase.J Biol Chem. 1999 Oct 8;274(41):29433-8. doi: 10.1074/jbc.274.41.29433. J Biol Chem. 1999. PMID: 10506206
-
Three amino acid substitutions in domain I of calmodulin prevent the activation of chicken smooth muscle myosin light chain kinase.J Biol Chem. 1991 Nov 15;266(32):21488-95. J Biol Chem. 1991. PMID: 1657969
-
Myosin light chain kinase as a multifunctional regulatory protein of smooth muscle contraction.IUBMB Life. 2001 Jun;51(6):337-44. doi: 10.1080/152165401753366087. IUBMB Life. 2001. PMID: 11758800 Review.
Cited by
-
The dominant protein phosphatase PP1c isoform in smooth muscle cells, PP1cβ, is essential for smooth muscle contraction.J Biol Chem. 2018 Oct 26;293(43):16677-16686. doi: 10.1074/jbc.RA118.003083. Epub 2018 Sep 5. J Biol Chem. 2018. PMID: 30185619 Free PMC article.
-
Kinetics of myosin light chain kinase activation of smooth muscle myosin in an in vitro model system.Biochemistry. 2013 Nov 26;52(47):8489-500. doi: 10.1021/bi401001x. Epub 2013 Nov 11. Biochemistry. 2013. PMID: 24144337 Free PMC article.
-
Diffusion of myosin light chain kinase on actin: A mechanism to enhance myosin phosphorylation rates in smooth muscle.J Gen Physiol. 2015 Oct;146(4):267-80. doi: 10.1085/jgp.201511483. J Gen Physiol. 2015. PMID: 26415568 Free PMC article.
-
Increasing evidence of mechanical force as a functional regulator in smooth muscle myosin light chain kinase.Elife. 2017 Jul 11;6:e26473. doi: 10.7554/eLife.26473. Elife. 2017. PMID: 28696205 Free PMC article.
-
A role for the Ca(2+)-dependent tyrosine kinase Pyk2 in tonic depolarization-induced vascular smooth muscle contraction.J Muscle Res Cell Motil. 2015 Dec;36(6):479-89. doi: 10.1007/s10974-015-9416-2. Epub 2015 Jul 7. J Muscle Res Cell Motil. 2015. PMID: 26150074 Review.
References
-
- Walsh MP, Dabrowska R, Hinkins S, Hartshorne DJ. Calcium-independent myosin light chain kinase of smooth muscle. Preparation by limited chymotryptic digestion of the calcium ion dependent enzyme, purification, and characterization. Biochemistry. 1982;21:1919–1925. - PubMed
-
- Mayr GW, Heilmeyer LM., Jr Skeletal muscle myosin light chain kinase. A refined structural model. FEBS Lett. 1983;157:225–231. - PubMed
-
- Taylor SS, Knighton DR, Zheng J, Ten Eyck LF, Sowadski JM. Structural framework for the protein kinase family. Annu Rev Cell Biol. 1992;8:429–462. - PubMed
-
- Pearson RB, Wettenhall RE, Means AR, Hartshorne DJ, Kemp BE. Autoregulation of enzymes by pseudosubstrate prototopes: myosin light chain kinase. Science. 1988;241:970–973. - PubMed
-
- Kemp BE, Pearson RB, House C, Robinson PJ, Means AR. Regulation of protein kinases by pseudosubstrate prototopes. Cell Sign. 1989;1:303–311. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

