Probing the DNA-binding affinity and specificity of designed zinc finger proteins
- PMID: 20197039
- PMCID: PMC2830443
- DOI: 10.1016/j.bpj.2009.11.021
Probing the DNA-binding affinity and specificity of designed zinc finger proteins
Abstract
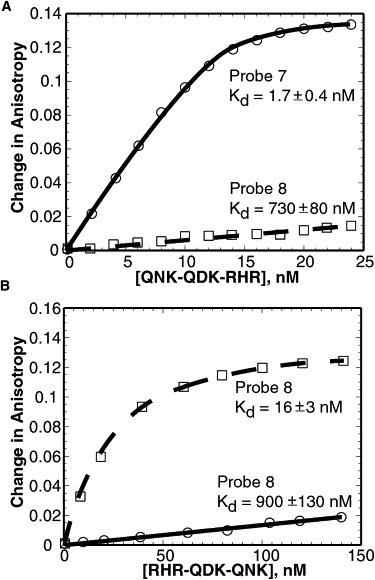
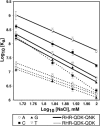
Engineered transcription factors and endonucleases based on designed Cys(2)His(2) zinc finger domains have proven to be effective tools for the directed regulation and modification of genes. The introduction of this technology into both research and clinical settings necessitates the development of rapid and accurate means of evaluating both the binding affinity and binding specificity of designed zinc finger domains. Using a fluorescence anisotropy-based DNA-binding assay, we examined the DNA-binding properties of two engineered zinc finger proteins that differ by a single amino acid. We demonstrate that the protein with the highest affinity for a particular DNA site need not be the protein that binds that site with the highest degree of specificity. Moreover, by comparing the binding characteristics of the two proteins at varying salt concentrations, we show that the ionic strength makes significant and variable contributions to both affinity and specificity. These results have significant implications for zinc finger design as they highlight the importance of considering affinity, specificity, and environmental requirements in designing a DNA-binding domain for a particular application.
2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Insights into the molecular recognition of the 5'-GNN-3' family of DNA sequences by zinc finger domains.J Mol Biol. 2000 Nov 3;303(4):489-502. doi: 10.1006/jmbi.2000.4133. J Mol Biol. 2000. PMID: 11054286
-
Assessment of major and minor groove DNA interactions by the zinc fingers of Xenopus transcription factor IIIA.Nucleic Acids Res. 1996 Jul 1;24(13):2567-74. doi: 10.1093/nar/24.13.2567. Nucleic Acids Res. 1996. PMID: 8692697 Free PMC article.
-
DNA-induced alpha-helix capping in conserved linker sequences is a determinant of binding affinity in Cys(2)-His(2) zinc fingers.J Mol Biol. 2000 Jan 28;295(4):719-27. doi: 10.1006/jmbi.1999.3406. J Mol Biol. 2000. PMID: 10656784
-
Engineering and Application of Zinc Finger Proteins and TALEs for Biomedical Research.Mol Cells. 2017 Aug;40(8):533-541. doi: 10.14348/molcells.2017.0139. Epub 2017 Aug 23. Mol Cells. 2017. PMID: 28835021 Free PMC article. Review.
-
Keep your fingers off my DNA: protein-protein interactions mediated by C2H2 zinc finger domains.Cell Biochem Biophys. 2008;50(3):111-31. doi: 10.1007/s12013-008-9008-5. Epub 2008 Feb 6. Cell Biochem Biophys. 2008. PMID: 18253864 Review.
Cited by
-
Complex signal processing in synthetic gene circuits using cooperative regulatory assemblies.Science. 2019 May 10;364(6440):593-597. doi: 10.1126/science.aau8287. Epub 2019 Apr 18. Science. 2019. PMID: 31000590 Free PMC article.
-
Direct Transcriptional Effects of Apolipoprotein E.J Neurosci. 2016 Jan 20;36(3):685-700. doi: 10.1523/JNEUROSCI.3562-15.2016. J Neurosci. 2016. PMID: 26791201 Free PMC article.
-
Zinc finger structure-function in Ikaros Marvin A Payne.World J Biol Chem. 2011 Jun 26;2(6):161-6. doi: 10.4331/wjbc.v2.i6.161. World J Biol Chem. 2011. PMID: 21765982 Free PMC article.
-
Leukocyte protease binding to nucleic acids promotes nuclear localization and cleavage of nucleic acid binding proteins.J Immunol. 2014 Jun 1;192(11):5390-7. doi: 10.4049/jimmunol.1303296. Epub 2014 Apr 25. J Immunol. 2014. PMID: 24771851 Free PMC article. Clinical Trial.
-
Modulating gene regulation to treat genetic disorders.Nat Rev Drug Discov. 2020 Nov;19(11):757-775. doi: 10.1038/s41573-020-0083-7. Epub 2020 Oct 5. Nat Rev Drug Discov. 2020. PMID: 33020616 Review.
References
-
- Beerli R.R., Barbas C.F. Engineering polydactyl zinc-finger transcription factors. Nat. Biotechnol. 2002;20:135–141. - PubMed
-
- Jantz D., Amann B.T., Berg J.M. The design of functional DNA-binding proteins based on zinc finger domains. Chem. Rev. 2004;104:789–799. - PubMed
-
- Klug A. Zinc finger peptides for the regulation of gene expression. J. Mol. Biol. 1999;293:215–218. - PubMed
-
- Pabo C.O., Peisach E., Grant R.A. Design and selection of novel Cys2His2 zinc finger proteins. Annu. Rev. Biochem. 2001;70:313–340. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

