Statistics and physical origins of pK and ionization state changes upon protein-ligand binding
- PMID: 20197041
- PMCID: PMC2830434
- DOI: 10.1016/j.bpj.2009.11.016
Statistics and physical origins of pK and ionization state changes upon protein-ligand binding
Abstract
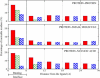
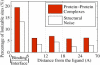
This work investigates statistical prevalence and overall physical origins of changes in charge states of receptor proteins upon ligand binding. These changes are explored as a function of the ligand type (small molecule, protein, and nucleic acid), and distance from the binding region. Standard continuum solvent methodology is used to compute, on an equal footing, pK changes upon ligand binding for a total of 5899 ionizable residues in 20 protein-protein, 20 protein-small molecule, and 20 protein-nucleic acid high-resolution complexes. The size of the data set combined with an extensive error and sensitivity analysis allows us to make statistically justified and conservative conclusions: in 60% of all protein-small molecule, 90% of all protein-protein, and 85% of all protein-nucleic acid complexes there exists at least one ionizable residue that changes its charge state upon ligand binding at physiological conditions (pH = 6.5). Considering the most biologically relevant pH range of 4-8, the number of ionizable residues that experience substantial pK changes (DeltapK > 1.0) due to ligand binding is appreciable: on average, 6% of all ionizable residues in protein-small molecule complexes, 9% in protein-protein, and 12% in protein-nucleic acid complexes experience a substantial pK change upon ligand binding. These changes are safely above the statistical false-positive noise level. Most of the changes occur in the immediate binding interface region, where approximately one out of five ionizable residues experiences substantial pK change regardless of the ligand type. However, the physical origins of the change differ between the types: in protein-nucleic acid complexes, the pK values of interface residues are predominantly affected by electrostatic effects, whereas in protein-protein and protein-small molecule complexes, structural changes due to the induced-fit effect play an equally important role. In protein-protein and protein-nucleic acid complexes, there is a statistically significant number of substantial pK perturbations, mostly due to the induced-fit structural changes, in regions far from the binding interface.
2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
References
-
- Voet D., Voet J.G. 2nd Ed. John Wiley & Sons; New York: 1995. Biochemistry. - PubMed
-
- de Silva A.P., Gunaratne H.Q., Rice T.E. Signaling recognition events with fluorescent sensors and switches. Chem. Rev. 1997;97:1515–1566. - PubMed
-
- Jorgensen W.L. The many roles of computation in drug discovery. Science. 2004;303:1813–1818. - PubMed
-
- Goh C.-S., Milburn D., Gerstein M. Conformational changes associated with protein-protein interactions. Curr. Opin. Struct. Biol. 2004;14:104–109. - PubMed
-
- Holmes K.C., Angert I., Schröder R.R. Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature. 2003;425:423–427. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

