Engineered interfaces of an AAA+ ATPase reveal a new nucleotide-dependent coordination mechanism
- PMID: 20197281
- PMCID: PMC2865273
- DOI: 10.1074/jbc.M110.103150
Engineered interfaces of an AAA+ ATPase reveal a new nucleotide-dependent coordination mechanism
Abstract
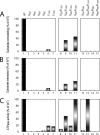
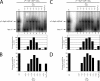
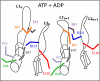
Homohexameric ring AAA(+) ATPases are found in all kingdoms of life and are involved in all cellular processes. To accommodate the large spectrum of substrates, the conserved AAA(+) core has become specialized through the insertion of specific substrate-binding motifs. Given their critical roles in cellular function, understanding the nucleotide-driven mechanisms of action is of wide importance. For one type of member AAA(+) protein (phage shock protein F, PspF), we identified and established the functional significance of strategically placed arginine and glutamate residues that form interacting pairs in response to nucleotide binding. We show that these interactions are critical for "cis" and "trans" subunit communication, which support coordination between subunits for nucleotide-dependent substrate remodeling. Using an allele-specific suppression approach for ATPase and substrate remodeling, we demonstrate that the targeted residues directly interact and are unlikely to make any other pairwise critical interactions. We then propose a mechanistic rationale by which the nucleotide-bound state of adjacent subunits can be sensed without direct involvement of R-finger residues. As the structural AAA(+) core is conserved, we propose that the functional networks established here could serve as a template to identify similar residue pairs in other AAA(+) proteins.
Figures
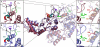


References
-
- Konakova M., Pulst S. M. (2005) J. Mol. Neurosci. 25, 105–117 - PubMed
-
- Guinto J. B., Ritson G. P., Taylor J. P., Forman M. S. (2007) Acta Neuropathol. 114, 55–61 - PubMed
-
- Kimonis V. E., Watts G. D. (2005) Alzheimer Dis. Assoc. Disord. 19, S44–S47 - PubMed
-
- Watts G. D., Mehta S. G., Zhao C., Ramdeen S., Hamilton S. J., Novack D. V., Mumm S., Whyte M. P., Mc Gillivray B., Kimonis V. E. (2005) Hum. Genet. 118, 508–514 - PubMed
-
- Watts G. D., Thomasova D., Ramdeen S. K., Fulchiero E. C., Mehta S. G., Drachman D. A., Weihl C. C., Jamrozik Z., Kwiecinski H., Kaminska A., Kimonis V. E. (2007) Clin. Genet. 72, 420–426 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

