SOMBRERO, BEARSKIN1, and BEARSKIN2 regulate root cap maturation in Arabidopsis
- PMID: 20197506
- PMCID: PMC2861445
- DOI: 10.1105/tpc.109.072272
SOMBRERO, BEARSKIN1, and BEARSKIN2 regulate root cap maturation in Arabidopsis
Abstract
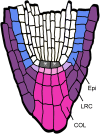
The root cap has a central role in root growth, determining the growth trajectory and facilitating penetration into the soil. Root cap cells have specialized functions and morphologies, and border cells are released into the rhizosphere by specific cell wall modifications. Here, we demonstrate that the cellular maturation of root cap is redundantly regulated by three genes, SOMBRERO (SMB), BEARSKIN1 (BRN1), and BRN2, which are members of the Class IIB NAC transcription factor family, together with the VASCULAR NAC DOMAIN (VND) and NAC SECONDARY WALL THICKENING PROMOTING FACTOR (NST) genes that regulate secondary cell wall synthesis in specialized cell types. Lateral cap cells in smb-3 mutants continue to divide and fail to detach from the root, phenotypes that are independent of FEZ upregulation in smb-3. In brn1-1 brn2-1 double mutants, columella cells fail to detach, while in triple mutants, cells fail to mature in all parts of the cap. This complex genetic redundancy involves differences in expression, protein activity, and target specificity. All three genes have very similar overexpression phenotypes to the VND/NST genes, indicating that members of this family are largely functionally equivalent. Our results suggest that Class IIB NAC proteins regulate cell maturation in cells that undergo terminal differentiation with strong cell wall modifications.
Figures






References
-
- Abas L., Benjamins R., Malenica N., Paciorek T., Wiśniewska J., Moulinier-Anzola J.C., Sieberer T., Friml J., Luschnig C. (2006). Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 8: 249–256 - PubMed
-
- Barlow P.W. (2003). The root cap: Cell dynamics, cell differentiation and cap function. J. Plant Growth Regul. 21: 261–286
-
- Bengough A.G., McKenzie B.M. (1997). Sloughing of root cap cells decreases the frictional resistance to maize (Zea mays L.) root growth. J. Exp. Bot. 48: 885–893
-
- Birnbaum K., Shasha D.E., Wang J.Y., Jung J.W., Lambert G.M., Galbraith D.W., Benfey P.N. (2003). A gene expression map of the Arabidopsis root. Science 302: 1956–1960 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

