CD99 inhibits neural differentiation of human Ewing sarcoma cells and thereby contributes to oncogenesis
- PMID: 20197622
- PMCID: PMC2827943
- DOI: 10.1172/JCI36667
CD99 inhibits neural differentiation of human Ewing sarcoma cells and thereby contributes to oncogenesis
Abstract
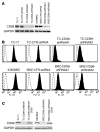
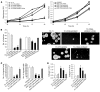
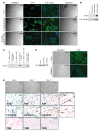
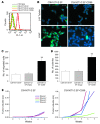
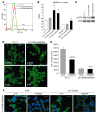
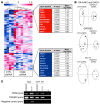
Ewing sarcoma (EWS) is an aggressive bone tumor of uncertain cellular origin. CD99 is a membrane protein that is expressed in most cases of EWS, although its function in the disease is unknown. Here we have shown that endogenous CD99 expression modulates EWS tumor differentiation and malignancy. We determined that knocking down CD99 expression in human EWS cell lines reduced their ability to form tumors and bone metastases when xenografted into immunodeficient mice and diminished their tumorigenic characteristics in vitro. Further, reduction of CD99 expression resulted in neurite outgrowth and increased expression of beta-III tubulin and markers of neural differentiation. Analysis of a panel of human EWS cells revealed an inverse correlation between CD99 and H-neurofilament expression, as well as an inverse correlation between neural differentiation and oncogenic transformation. As knockdown of CD99 also led to an increase in phosphorylation of ERK1/2, we suggest that the CD99-mediated prevention of neural differentiation of EWS occurs through MAPK pathway modulation. Together, these data indicate a new role for CD99 in preventing neural differentiation of EWS cells and suggest that blockade of CD99 or its downstream molecular pathway may be a new therapeutic approach for EWS.
Figures

Similar articles
-
MiR-30a-5p connects EWS-FLI1 and CD99, two major therapeutic targets in Ewing tumor.Oncogene. 2013 Aug 15;32(33):3915-21. doi: 10.1038/onc.2012.403. Oncogene. 2013. PMID: 22986530
-
CD99 triggering in Ewing sarcoma delivers a lethal signal through p53 pathway reactivation and cooperates with doxorubicin.Clin Cancer Res. 2015 Jan 1;21(1):146-56. doi: 10.1158/1078-0432.CCR-14-0492. Epub 2014 Dec 11. Clin Cancer Res. 2015. PMID: 25501132
-
Exosomes from CD99-deprived Ewing sarcoma cells reverse tumor malignancy by inhibiting cell migration and promoting neural differentiation.Cell Death Dis. 2019 Jun 17;10(7):471. doi: 10.1038/s41419-019-1675-1. Cell Death Dis. 2019. PMID: 31209202 Free PMC article.
-
Ewing sarcoma family of tumors.Adv Anat Pathol. 2005 Jul;12(4):212-20. doi: 10.1097/01.pap.0000175114.55541.52. Adv Anat Pathol. 2005. PMID: 16096383 Review.
-
Ewing Sarcoma, an enigmatic malignancy of likely progenitor cell origin, driven by transcription factor oncogenic fusions.Int J Clin Exp Pathol. 2010 Mar 19;3(4):338-47. Int J Clin Exp Pathol. 2010. PMID: 20490326 Free PMC article. Review.
Cited by
-
Primary intradural extramedullary Ewing sarcoma in the lumbar area: A case report.Radiol Case Rep. 2022 Sep 29;17(12):4617-4621. doi: 10.1016/j.radcr.2022.09.033. eCollection 2022 Dec. Radiol Case Rep. 2022. PMID: 36204406 Free PMC article.
-
Development of an Ewing sarcoma cell line with resistance to EWS‑FLI1 inhibitor YK‑4‑279.Mol Med Rep. 2020 Mar;21(3):1667-1675. doi: 10.3892/mmr.2020.10948. Epub 2020 Jan 16. Mol Med Rep. 2020. PMID: 32016454 Free PMC article.
-
CD99 triggering induces methuosis of Ewing sarcoma cells through IGF-1R/RAS/Rac1 signaling.Oncotarget. 2016 Nov 29;7(48):79925-79942. doi: 10.18632/oncotarget.13160. Oncotarget. 2016. PMID: 27835596 Free PMC article.
-
Targeting Histone Deacetylase Activity to Arrest Cell Growth and Promote Neural Differentiation in Ewing Sarcoma.Mol Neurobiol. 2018 Sep;55(9):7242-7258. doi: 10.1007/s12035-018-0874-6. Epub 2018 Feb 3. Mol Neurobiol. 2018. PMID: 29397557 Free PMC article.
-
Targeted therapies for advanced Ewing sarcoma family of tumors.Cancer Treat Rev. 2015 May;41(5):391-400. doi: 10.1016/j.ctrv.2015.03.008. Epub 2015 Mar 27. Cancer Treat Rev. 2015. PMID: 25869102 Free PMC article. Review.
References
-
- Bernstein M, et al. Ewing’s sarcoma family of tumors: current management. Oncologist. 2006;11(5):503–519. - PubMed
-
- Henderson DW, Leppard PJ, Brennan JS, Mukherjee TM, Swift JG. Primitive neuroepithelial tumours of soft tissues and of bone: further ultrastructural and immunocytochemical clarification of ‘Ewing’s sarcoma’, including freeze-fracture analysis. J Submicrosc Cytol Pathol. 1989;21(1):35–57. - PubMed
-
- Lipinski M, et al. Neuroectoderm-associated antigens on Ewing’s sarcoma cell lines. Cancer Res. 1987;47(1):183–187. - PubMed
-
- Lizard-Nacol S, Volk C, Lizard G, Turc-Carel C. Abnormal expression of neurofilament proteins in Ewing’s sarcoma cell cultures. Tumour Biol. 1992;13(1–2):36–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

