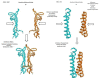
Types of interfaces for homodimer folding and binding
- PMID: 20198182
- PMCID: PMC2828894
- DOI: 10.6026/97320630007101
Types of interfaces for homodimer folding and binding
Abstract
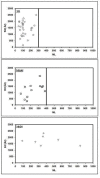
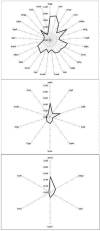
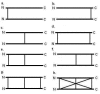
Homodimers have a role in catalysis and regulation through the formation of stable interfaces. These interfaces are formed through different folding mechanisms such as 2-state without stable intermediate (2S), 3-state with monomer intermediate (3SMI) and 3-state with dimer intermediate (3SDI). Therefore, it is of interest to understand folding mechanism using structural features at the interfaces. Several studies have documented the significance of structural features for the understanding of homodimer folding mechanisms. However, the known features provide limited information for understanding homodimer folding mechanisms. Hence, we created an extended dataset of 47 homodimers (twenty eight 2S, twelve 3SMI and seven 3SDI) to examine the types of interfaces in protein homodimers. 2S are usually small sized, 3SMI are often medium sized and 3SDI often exist as large sized proteins. The ratio of interface to total (I/T) residue is large in 2S and small in 3SMI and 3SDI. Hence, we used I/T measure to group 2S, 3SMI and 3SDI into categories with large I/T (>> 50%), moderate I/T (50 - 25%) and small I/T (<< 25%) interfaces. The grouping is further sub-grouped based on the type of physical interaction visualized at the interface using representations in two dimensions (2D). 2D representation of the interface shows eight different forms of interactions in these homodimers. 2S homodimers frequently have large I/T and thus, utilize the entire protein structure in the formation of the interface where the individual subunits are heavily inter communicated with each other. This is not true in the case of 3SMI and 3SDI. 3SMI subunits usually interact with each other at the interface with a gentle touch-like contact and hence, they have low I/T ratio. 3SDI are often quite different in interaction compared to 3SMI and their subunits do deeply interact at the interface with only one part of the surface and hence also having low I/T ratio.
Keywords: homodimer; interaction; interface; mode; structures; types.
Figures





Similar articles
-
Structural features for homodimer folding mechanism.J Mol Graph Model. 2009 Sep;28(2):88-94. doi: 10.1016/j.jmgm.2009.04.002. Epub 2009 Apr 19. J Mol Graph Model. 2009. PMID: 19442545
-
A decision tree model for the prediction of homodimer folding mechanism.Bioinformation. 2009 Nov 17;4(5):197-205. doi: 10.6026/97320630004197. Bioinformation. 2009. PMID: 20461159 Free PMC article.
-
Structural features differentiate the mechanisms between 2S (2 state) and 3S (3 state) folding homodimers.Bioinformation. 2005 Sep 2;1(2):42-9. doi: 10.6026/97320630001042. Bioinformation. 2005. PMID: 17597851 Free PMC article.
-
Structural motifs at protein-protein interfaces: protein cores versus two-state and three-state model complexes.Protein Sci. 1997 Sep;6(9):1793-805. doi: 10.1002/pro.5560060901. Protein Sci. 1997. PMID: 9300480 Free PMC article. Review.
-
Inter-residue interactions in protein folding and stability.Prog Biophys Mol Biol. 2004 Oct;86(2):235-77. doi: 10.1016/j.pbiomolbio.2003.09.003. Prog Biophys Mol Biol. 2004. PMID: 15288760 Review.
Cited by
-
From Anna University to America and to Agriculture.Bioinformation. 2021 Jan 31;17(1):29-36. doi: 10.6026/97320630017029. eCollection 2021. Bioinformation. 2021. PMID: 34393415 Free PMC article.
-
Improving accuracy of protein contact prediction using balanced network deconvolution.Proteins. 2015 Mar;83(3):485-96. doi: 10.1002/prot.24744. Epub 2015 Jan 24. Proteins. 2015. PMID: 25524593 Free PMC article.
-
Whole-genome re-sequencing reveals the impact of the interaction of copy number variants of the rhg1 and Rhg4 genes on broad-based resistance to soybean cyst nematode.Plant Biotechnol J. 2019 Aug;17(8):1595-1611. doi: 10.1111/pbi.13086. Epub 2019 Feb 20. Plant Biotechnol J. 2019. PMID: 30688400 Free PMC article.
-
Geometrical and electro-static determinants of protein-protein interactions.Bioinformation. 2021 Oct 31;17(10):851-860. doi: 10.6026/97320630017851. eCollection 2021. Bioinformation. 2021. PMID: 35574504 Free PMC article.
References
LinkOut - more resources
Full Text Sources
