A docking model of human ribonucleotide reductase with flavin and phenosafranine
- PMID: 20198185
- PMCID: PMC2828892
- DOI: 10.6026/97320630004123
A docking model of human ribonucleotide reductase with flavin and phenosafranine
Abstract
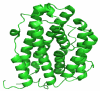
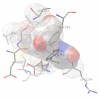
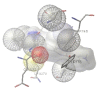
Ribonucleotide Reductase (RNR) is an enzyme responsible for the reduction of ribonucleotides to their corresponding Deoxyribonucleotides (DNA), which is a building block for DNA replication and repair mechanisms. The key role of RNR in DNA synthesis and control in cell growth has made this an important target for anticancer therapy. Increased RNR activity has been associated with malignant transformation and tumor cell growth. In recent years, several RNR inhibitors, including Triapine, Gemcitabine and GTI-2040, have entered the clinical trials. Our current work focuses on an attempted to dock this inhibitors Flavin and Phenosafranine to curtail the action of human RNR2. The docked inhibitor Flavin and Phenosafranine binds at the active site with THR176, which are essential for free radical formation. The inhibitor must be a radical scavenger to destroy the tyrosyl radical or iron metal scavenger. The iron or radical site of R2 protein can react with one-electron reductants, whereby the tyrosyl radical is converted to a normal tyrosine residue. However, compounds such as Flavin and Phenosafranine were used in most of the cases to reduce the radical activity. The docking study was performed for the crystal structure of human RNR with the radical scavengers Flavin and Phenosafranine to inhibit the human RNR2. This helps to understand the functional aspects and also aids in the development of novel inhibitors for the human RNR2.
Keywords: Flavin; Phenosafranine; Ribonucleotide reductase; inhibitors; radical scavenger.
Figures
Similar articles
-
Modeling and Proposed Molecular Mechanism of Hydroxyurea Through Docking and Molecular Dynamic Simulation to Curtail the Action of Ribonucleotide Reductase.Recent Pat Anticancer Drug Discov. 2016;11(4):461-468. doi: 10.2174/1574892811666160926143534. Recent Pat Anticancer Drug Discov. 2016. PMID: 27670694
-
Structure, function, and mechanism of ribonucleotide reductases.Biochim Biophys Acta. 2004 Jun 1;1699(1-2):1-34. doi: 10.1016/j.bbapap.2004.02.007. Biochim Biophys Acta. 2004. PMID: 15158709 Review.
-
Ribonucleotide reductase: Implications of thiol S-nitrosylation and tyrosine nitration for different subunits.Nitric Oxide. 2022 Oct 1;127:26-43. doi: 10.1016/j.niox.2022.07.002. Epub 2022 Jul 15. Nitric Oxide. 2022. PMID: 35850377 Review.
-
Metal-free ribonucleotide reduction powered by a DOPA radical in Mycoplasma pathogens.Nature. 2018 Nov;563(7731):416-420. doi: 10.1038/s41586-018-0653-6. Epub 2018 Oct 31. Nature. 2018. PMID: 30429545 Free PMC article.
-
Spectroscopic studies of the iron and manganese reconstituted tyrosyl radical in Bacillus cereus ribonucleotide reductase R2 protein.PLoS One. 2012;7(3):e33436. doi: 10.1371/journal.pone.0033436. Epub 2012 Mar 14. PLoS One. 2012. PMID: 22432022 Free PMC article.
Cited by
-
Spectroscopic characterization of the interaction of phenosafranin and safranin O with double stranded, heat denatured and single stranded calf thymus DNA.J Fluoresc. 2011 Jan;21(1):247-55. doi: 10.1007/s10895-010-0712-3. Epub 2010 Sep 28. J Fluoresc. 2011. PMID: 20878351
-
How Does Nanoconfinement within a Reverse Micelle Influence the Interaction of Phenazinium-Based Photosensitizers with DNA?ACS Omega. 2018 Feb 1;3(2):1374-1385. doi: 10.1021/acsomega.7b01820. eCollection 2018 Feb 28. ACS Omega. 2018. PMID: 31458466 Free PMC article.
-
Cationic Phenosafranin Photosensitizers Based on Polyhedral Oligomeric Silsesquioxanes for Inactivation of Gram-Positive and Gram-Negative Bacteria.Int J Mol Sci. 2021 Dec 13;22(24):13373. doi: 10.3390/ijms222413373. Int J Mol Sci. 2021. PMID: 34948170 Free PMC article.
References
LinkOut - more resources
Full Text Sources
