CARON--average RMSD of NMR structure ensembles
- PMID: 20198187
- PMCID: PMC2828896
- DOI: 10.6026/97320630004132
CARON--average RMSD of NMR structure ensembles
Abstract
The NMR protein structures are often deposited in the Protein Data Bank as ensembles of models that agree with the experimental restraints. Information about stereochemical variability and the molecular flexibility can be obtained by systematic comparison of all models. Here we describe CARON, a software that allows the computation of the root-mean-square-distances between equivalent atoms and residues in all models and introduces these values into the occupancy and the B-factor fields of PDB-formatted files. This tool allows the user to both get a quantitative estimation of the conformational homogeneity of the models and to exploit this information in common computer graphics programs.
Keywords: Bioinformatics software; CARON; NMR spectroscopy; Protein Data Bank; Root-Mean-Square Deviation; conformational homogeneity; superposition.
Figures
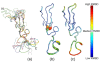
Trace of all 10 models of the 1HRF entry of the Protein Data Bank;
First model colored according to the B-factor putty script distributed with PyMol;
First model colored according to the average residues RMSD computed with CARON.
References
-
- Bernstein FC, et al. J Mol Biol. 1977;112:535. - PubMed
-
- Ying-Hung L, et al. International Computer Symposium (ICS'2004); Taipei, Taiwan. 2004. pp. 1000–1005.
-
- Kabsch W. Acta Crystallographica. 1976;32:922.
LinkOut - more resources
Full Text Sources
