Use of the growing environment as a source of variation to identify the quantitative trait transcripts and modules of co-expressed genes that determine chlorogenic acid accumulation
- PMID: 20199615
- PMCID: PMC2904492
- DOI: 10.1111/j.1365-3040.2010.02141.x
Use of the growing environment as a source of variation to identify the quantitative trait transcripts and modules of co-expressed genes that determine chlorogenic acid accumulation
Abstract
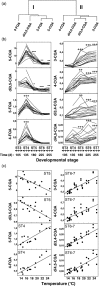
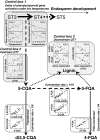
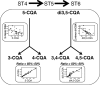
Developing Coffea arabica seeds accumulate large amounts of chlorogenic acids (CGAs) as a storage form of phenylpropanoid derivatives, making coffee a valuable model to investigate the metabolism of these widespread plant phenolics. However, developmental and environmental regulations of CGA metabolism are poorly understood. In the present work, the expression of selected phenylpropanoid genes, together with CGA isomer profiles, was monitored throughout seed development across a wide set of contrasted natural environments. Although CGA metabolism was controlled by major developmental factors, the mean temperature during seed development had a direct impact on the time-window of CGA biosynthesis, as well as on final CGA isomer composition through subtle transcriptional regulations. We provide evidence that the variability induced by the environment is a useful tool to test whether CGA accumulation is quantitatively modulated at the transcriptional level, hence enabling detection of rate-limiting transcriptional steps [quantitative trait transcripts (QTTs)] for CGA biosynthesis. Variations induced by the environment also enabled a better description of the phenylpropanoid gene transcriptional network throughout seed development, as well as the detection of three temporally distinct modules of quantitatively co-expressed genes. Finally, analysis of metabolite-to-metabolite relationships revealed new biochemical characteristics of the isomerization steps that remain uncharacterized at the gene level.
Figures


Similar articles
-
Metabolic pathways in tropical dicotyledonous albuminous seeds: Coffea arabica as a case study.New Phytol. 2009;182(1):146-162. doi: 10.1111/j.1469-8137.2008.02742.x. Epub 2009 Jan 21. New Phytol. 2009. PMID: 19207685 Free PMC article.
-
Regulation of galactomannan biosynthesis in coffee seeds.J Exp Bot. 2014 Jan;65(1):323-37. doi: 10.1093/jxb/ert380. Epub 2013 Nov 7. J Exp Bot. 2014. PMID: 24203356
-
Genetic mapping of a caffeoyl-coenzyme A 3-O-methyltransferase gene in coffee trees. Impact on chlorogenic acid content.Theor Appl Genet. 2003 Aug;107(4):751-6. doi: 10.1007/s00122-003-1310-4. Epub 2003 Jul 12. Theor Appl Genet. 2003. PMID: 12861362
-
Should I stay or should I go: are chlorogenic acids mobilized towards lignin biosynthesis?Phytochemistry. 2019 Oct;166:112063. doi: 10.1016/j.phytochem.2019.112063. Epub 2019 Jul 4. Phytochemistry. 2019. PMID: 31280091 Review.
-
Research Advances in the Synthesis, Metabolism, and Function of Chlorogenic Acid.Foods. 2025 May 28;14(11):1914. doi: 10.3390/foods14111914. Foods. 2025. PMID: 40509442 Free PMC article. Review.
Cited by
-
Exploiting the Genetic Diversity of Maize Using a Combined Metabolomic, Enzyme Activity Profiling, and Metabolic Modeling Approach to Link Leaf Physiology to Kernel Yield.Plant Cell. 2017 May;29(5):919-943. doi: 10.1105/tpc.16.00613. Epub 2017 Apr 10. Plant Cell. 2017. PMID: 28396554 Free PMC article.
-
Volatile Metabolite Profiles of Robusta Green Bean Coffee From Different Geographical Origins in West Java and Their Correlation With Temperature, Rainfall, and Altitudes Using SPME GC-MS-Based Metabolomics.Int J Food Sci. 2024 Oct 26;2024:6908059. doi: 10.1155/2024/6908059. eCollection 2024. Int J Food Sci. 2024. PMID: 39494365 Free PMC article.
-
Potential biomonitoring of atmospheric carbon dioxide in Coffea arabica leaves using near-infrared spectroscopy and partial least squares discriminant analysis.Environ Sci Pollut Res Int. 2019 Oct;26(29):30356-30364. doi: 10.1007/s11356-019-06163-1. Epub 2019 Aug 21. Environ Sci Pollut Res Int. 2019. PMID: 31432374
-
Machine learning and statistics to qualify environments through multi-traits in Coffea arabica.PLoS One. 2021 Jan 12;16(1):e0245298. doi: 10.1371/journal.pone.0245298. eCollection 2021. PLoS One. 2021. PMID: 33434204 Free PMC article.
-
Constitutively elevated salicylic acid levels alter photosynthesis and oxidative state but not growth in transgenic populus.Plant Cell. 2013 Jul;25(7):2714-30. doi: 10.1105/tpc.113.112839. Epub 2013 Jul 31. Plant Cell. 2013. PMID: 23903318 Free PMC article.
References
-
- Aerts RJ, Baumann TW. Distribution and utilization of chlorogenic acid in Coffea seedlings. Journal of Experimental Botany. 1994;45:497–503.
-
- Bate NJ, Orr J, Ni WT, Meromi A, Nadlerhassar T, Doerner PW, Dixon RA, Lamb CJ, Elkind Y. Quantitative relationship between phenylalanine ammonia-lyase levels and phenylpropanoid accumulation in transgenic tobacco identifies a rate-determining step in natural product synthesis. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:7608–7612. - PMC - PubMed
-
- Bertrand C, Noirot M, Doulbeau S, de Kochko A, Hamon S, Campa C. Chlorogenic acid content swap during fruit maturation in Coffea pseudozanguebariae– qualitative comparison with leaves. Plant Science. 2003;165:1355–1361.
-
- Blodner C, Goebel C, Feussner I, Gatz C, Polle A. Warm and cold parental reproductive environments affect seed properties, fitness, and cold responsiveness in Arabidopsis thaliana progenies. Plant, Cell & Environment. 2007;30:165–175. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

