Entamoeba histolytica Phosphoserine aminotransferase (EhPSAT): insights into the structure-function relationship
- PMID: 20199659
- PMCID: PMC2850911
- DOI: 10.1186/1756-0500-3-52
Entamoeba histolytica Phosphoserine aminotransferase (EhPSAT): insights into the structure-function relationship
Abstract
Background: Presence of phosphorylated Serine biosynthesis pathway upstream to the de novo cysteine biosynthesis pathway makes PSAT a crucial enzyme. Besides this, phoshoserine produced by the enzyme can also be taken up directly by cysteine synthase as a substrate. PSAT is a PLP dependent enzyme where the cofactor serves as an epicenter for functional catalysis with the active site architecture playing crucial role in optimum function of the enzyme.
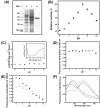
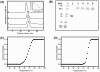
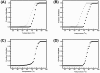
Findings: EhPSAT is a homodimer of molecular mass 86 kDa. To understand the structural modulations associated with pH dependent changes in functional activity of EhPSAT detailed biophysical studies were carried out. pH alterations had no significant effect on the secondary structure, cofactor orientation and oligomeric configuration of the enzyme however, pH dependent compaction in molecular dimensions was observed. Most interestingly, a direct correlation between pH induced modulation of functional activity and orientation of Trp 101 present in the active site of the enzyme was observed. Sodium halides nullified the pH induced global changes in the enzyme, however differential effect of these salts on the active site microenvironment and functional activity of the enzyme was observed.
Conclusions: The study unequivocally demonstrates that pH induced selective modification of active site microenvironment and not global change in structure or oligomeric status of the enzyme is responsible for the pH dependent change in enzymatic activity of PSAT.
Figures


Similar articles
-
Biophysical characterization of Entamoeba histolytica phosphoserine aminotransferase (EhPSAT): role of cofactor and domains in stability and subunit assembly.Eur Biophys J. 2011 May;40(5):599-610. doi: 10.1007/s00249-010-0654-3. Epub 2010 Dec 16. Eur Biophys J. 2011. PMID: 21161522
-
Biochemical and functional characterization of phosphoserine aminotransferase from Entamoeba histolytica, which possesses both phosphorylated and non-phosphorylated serine metabolic pathways.Mol Biochem Parasitol. 2006 Jan;145(1):71-83. doi: 10.1016/j.molbiopara.2005.09.008. Epub 2005 Oct 5. Mol Biochem Parasitol. 2006. PMID: 16289358
-
N-terminal residues are crucial for quaternary structure and active site conformation for the phosphoserine aminotransferase from enteric human parasite E. histolytica.Int J Biol Macromol. 2019 Jul 1;132:1012-1023. doi: 10.1016/j.ijbiomac.2019.04.027. Epub 2019 Apr 5. Int J Biol Macromol. 2019. PMID: 30959130
-
Phosphoserine Aminotransferase has Conserved Active Site from Microbes to Higher Eukaryotes with Minor Deviations.Protein Pept Lett. 2021;28(9):996-1008. doi: 10.2174/0929866528666210215140231. Protein Pept Lett. 2021. PMID: 33588715 Review.
-
Structure-Based Design of Inhibitors of the Crucial Cysteine Biosynthetic Pathway Enzyme O-Acetyl Serine Sulfhydrylase.Curr Top Med Chem. 2016;16(9):948-59. doi: 10.2174/1568026615666150825142422. Curr Top Med Chem. 2016. PMID: 26303427 Review.
Cited by
-
Molecular Structure of Phosphoserine Aminotransferase from Saccharomyces cerevisiae.Int J Mol Sci. 2023 Mar 7;24(6):5139. doi: 10.3390/ijms24065139. Int J Mol Sci. 2023. PMID: 36982214 Free PMC article.
-
Phenotypic and transcriptional profiling in Entamoeba histolytica reveal costs to fitness and adaptive responses associated with metronidazole resistance.Front Microbiol. 2015 May 5;6:354. doi: 10.3389/fmicb.2015.00354. eCollection 2015. Front Microbiol. 2015. PMID: 25999919 Free PMC article.
-
Identification of Small Molecule Inhibitors Targeting Phosphoserine Phosphatase: A Novel Target for the Development of Antiamoebic Drugs.ACS Omega. 2024 Jun 17;9(26):27906-27918. doi: 10.1021/acsomega.3c09439. eCollection 2024 Jul 2. ACS Omega. 2024. PMID: 38973836 Free PMC article.
-
L-serine biosynthesis in the human central nervous system: Structure and function of phosphoserine aminotransferase.Protein Sci. 2023 Apr;32(4):e4609. doi: 10.1002/pro.4609. Protein Sci. 2023. PMID: 36851825 Free PMC article.
-
Biophysical characterization of Entamoeba histolytica phosphoserine aminotransferase (EhPSAT): role of cofactor and domains in stability and subunit assembly.Eur Biophys J. 2011 May;40(5):599-610. doi: 10.1007/s00249-010-0654-3. Epub 2010 Dec 16. Eur Biophys J. 2011. PMID: 21161522
References
-
- Hester G, Stark W, Moser M, Kallen J, Markovic-Housley Z, Jansonius JN. Crystal structure of phosphoserine aminotransferase from Escherichia coli at 2.3 A resolution: comparison of the unligated enzyme and a complex with alpha-methyl-l-glutamate. J Mol Biol. 1999;286(3):829–850. doi: 10.1006/jmbi.1998.2506. - DOI - PubMed
-
- John RA. Pyridoxal phosphate-dependent enzymes. Biochim Biophys Acta. 1995;1248:81–96. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

