Conservation and divergence of known apicomplexan transcriptional regulons
- PMID: 20199665
- PMCID: PMC2841118
- DOI: 10.1186/1471-2164-11-147
Conservation and divergence of known apicomplexan transcriptional regulons
Abstract
Background: The apicomplexans are a diverse phylum of parasites causing an assortment of diseases including malaria in a wide variety of animals and lymphoproliferation in cattle. Little is known about how these varied parasites regulate their transcriptional regulons. Even less is known about how regulon systems, consisting of transcription factors and target genes together with their associated biological process, evolve in these diverse parasites.
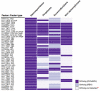
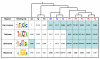
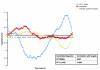
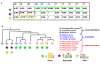
Results: In order to obtain insights into the differences in transcriptional regulation between these parasites we compared the orthology profiles of putative malaria transcription factors across species and examined the enrichment patterns of four binding sites across eleven apicomplexans. About three-fifths of the factors are broadly conserved in several phylogenetic orders of sequenced apicomplexans. This observation suggests the existence of regulons whose regulation is conserved across this ancient phylum. Transcription factors not broadly conserved across the phylum are possibly involved in regulon systems that have diverged between species. Examining binding site enrichment patterns in light of transcription factor conservation patterns suggests a second mode via which regulon systems may diverge - rewiring of existing transcription factors and their associated binding sites in specific ways. Integrating binding sites with transcription factor conservation patterns also facilitated prediction of putative regulators for one of the binding sites.
Conclusions: Even though transcription factors are underrepresented in apicomplexans, the distribution of these factors and their associated regulons reflect common and family-specific transcriptional regulatory processes.
Figures


Similar articles
-
The Cryptosporidium parvum ApiAP2 gene family: insights into the evolution of apicomplexan AP2 regulatory systems.Nucleic Acids Res. 2014 Jul;42(13):8271-84. doi: 10.1093/nar/gku500. Epub 2014 Jun 23. Nucleic Acids Res. 2014. PMID: 24957599 Free PMC article.
-
Horizontal gene transfer of epigenetic machinery and evolution of parasitism in the malaria parasite Plasmodium falciparum and other apicomplexans.BMC Evol Biol. 2013 Feb 11;13:37. doi: 10.1186/1471-2148-13-37. BMC Evol Biol. 2013. PMID: 23398820 Free PMC article.
-
Reconstruction of the core and extended regulons of global transcription factors.PLoS Genet. 2010 Jul 22;6(7):e1001027. doi: 10.1371/journal.pgen.1001027. PLoS Genet. 2010. PMID: 20661434 Free PMC article.
-
Phylogeny and evolution of apicoplasts and apicomplexan parasites.Parasitol Int. 2015 Jun;64(3):254-9. doi: 10.1016/j.parint.2014.10.005. Epub 2014 Oct 14. Parasitol Int. 2015. PMID: 25451217 Review.
-
Aspartyl proteinase genes from apicomplexan parasites: evidence for evolution of the gene structure.Trends Parasitol. 2001 Oct;17(10):491-8. doi: 10.1016/s1471-4922(01)02030-x. Trends Parasitol. 2001. PMID: 11587964 Review.
Cited by
-
Nascent RNA sequencing reveals mechanisms of gene regulation in the human malaria parasite Plasmodium falciparum.Nucleic Acids Res. 2017 Jul 27;45(13):7825-7840. doi: 10.1093/nar/gkx464. Nucleic Acids Res. 2017. PMID: 28531310 Free PMC article.
-
Red Blood Cell Invasion by the Malaria Parasite Is Coordinated by the PfAP2-I Transcription Factor.Cell Host Microbe. 2017 Jun 14;21(6):731-741.e10. doi: 10.1016/j.chom.2017.05.006. Cell Host Microbe. 2017. PMID: 28618269 Free PMC article.
-
The Cryptosporidium parvum ApiAP2 gene family: insights into the evolution of apicomplexan AP2 regulatory systems.Nucleic Acids Res. 2014 Jul;42(13):8271-84. doi: 10.1093/nar/gku500. Epub 2014 Jun 23. Nucleic Acids Res. 2014. PMID: 24957599 Free PMC article.
-
Cis regulatory motifs and antisense transcriptional control in the apicomplexan Theileria parva.BMC Genomics. 2016 Feb 20;17:128. doi: 10.1186/s12864-016-2444-5. BMC Genomics. 2016. PMID: 26896950 Free PMC article.
-
ApiAP2 Transcription Factors in Apicomplexan Parasites.Pathogens. 2019 Apr 7;8(2):47. doi: 10.3390/pathogens8020047. Pathogens. 2019. PMID: 30959972 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

