Genome-wide transcriptome analysis of the transition from primary to secondary stem development in Populus trichocarpa
- PMID: 20199690
- PMCID: PMC2846914
- DOI: 10.1186/1471-2164-11-150
Genome-wide transcriptome analysis of the transition from primary to secondary stem development in Populus trichocarpa
Abstract
Background: With its genome sequence and other experimental attributes, Populus trichocarpa has become the model species for genomic studies of wood development. Wood is derived from secondary growth of tree stems, and begins with the development of a ring of vascular cambium in the young developing stem. The terminal region of the developing shoot provides a steep developmental gradient from primary to secondary growth that facilitates identification of genes that play specialized functions during each of these phases of growth.
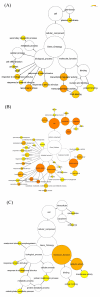
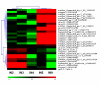
Results: Using a genomic microarray representing the majority of the transcriptome, we profiled gene expression in stem segments that spanned primary to secondary growth. We found 3,016 genes that were differentially expressed during stem development (Q-value </= 0.05; >2-fold expression variation), and 15% of these genes encode proteins with no significant identities to known genes. We identified all gene family members putatively involved in secondary growth for carbohydrate active enzymes, tubulins, actins, actin depolymerizing factors, fasciclin-like AGPs, and vascular development-associated transcription factors. Almost 70% of expressed transcription factors were upregulated during the transition to secondary growth. The primary shoot elongation region of the stem contained specific carbohydrate active enzyme and expansin family members that are likely to function in primary cell wall synthesis and modification. Genes involved in plant defense and protective functions were also dominant in the primary growth region.
Conclusion: Our results describe the global patterns of gene expression that occur during the transition from primary to secondary stem growth. We were able to identify three major patterns of gene expression and over-represented gene ontology categories during stem development. The new regulatory factors and cell wall biogenesis genes that we identified provide candidate genes for further functional characterization, as well as new tools for molecular breeding and biotechnology aimed at improvement of tree growth rate, crown form, and wood quality.
Figures









Similar articles
-
Identification and characterization of the Populus trichocarpa CLE family.BMC Genomics. 2016 Mar 2;17:174. doi: 10.1186/s12864-016-2504-x. BMC Genomics. 2016. PMID: 26935217 Free PMC article.
-
Genome-wide characterization of aspartic protease (AP) gene family in Populus trichocarpa and identification of the potential PtAPs involved in wood formation.BMC Plant Biol. 2019 Jun 24;19(1):276. doi: 10.1186/s12870-019-1865-0. BMC Plant Biol. 2019. PMID: 31234799 Free PMC article.
-
Validation of Reference Genes for Gene Expression by Quantitative Real-Time RT-PCR in Stem Segments Spanning Primary to Secondary Growth in Populus tomentosa.PLoS One. 2016 Jun 14;11(6):e0157370. doi: 10.1371/journal.pone.0157370. eCollection 2016. PLoS One. 2016. PMID: 27300480 Free PMC article.
-
Modeling transcriptional networks regulating secondary growth and wood formation in forest trees.Physiol Plant. 2014 Jun;151(2):156-63. doi: 10.1111/ppl.12113. Epub 2013 Oct 21. Physiol Plant. 2014. PMID: 24117954 Review.
-
Functional genomics of wood quality and properties.Genomics Proteomics Bioinformatics. 2003 Nov;1(4):263-78. doi: 10.1016/s1672-0229(03)01032-5. Genomics Proteomics Bioinformatics. 2003. PMID: 15629055 Free PMC article. Review.
Cited by
-
Discovery of genes that positively affect biomass and stress associated traits in poplar.Front Plant Sci. 2024 Oct 18;15:1468905. doi: 10.3389/fpls.2024.1468905. eCollection 2024. Front Plant Sci. 2024. PMID: 39494052 Free PMC article.
-
Genome-wide identification, evolutionary expansion, and expression profile of homeodomain-leucine zipper gene family in poplar (Populus trichocarpa).PLoS One. 2012;7(2):e31149. doi: 10.1371/journal.pone.0031149. Epub 2012 Feb 16. PLoS One. 2012. PMID: 22359569 Free PMC article.
-
Functional Diversification of Populus FLOWERING LOCUS D-LIKE3 Transcription Factor and Two Paralogs in Shoot Ontogeny, Flowering, and Vegetative Phenology.Front Plant Sci. 2022 Feb 3;13:805101. doi: 10.3389/fpls.2022.805101. eCollection 2022. Front Plant Sci. 2022. PMID: 35185983 Free PMC article.
-
Expression profiles of cell-wall related genes vary broadly between two common maize inbreds during stem development.BMC Genomics. 2019 Oct 29;20(1):785. doi: 10.1186/s12864-019-6117-z. BMC Genomics. 2019. PMID: 31664907 Free PMC article.
-
Next-generation sequencing-based mRNA and microRNA expression profiling analysis revealed pathways involved in the rapid growth of developing culms in Moso bamboo.BMC Plant Biol. 2013 Aug 21;13:119. doi: 10.1186/1471-2229-13-119. BMC Plant Biol. 2013. PMID: 23964682 Free PMC article.
References
-
- Jansson S, Douglas CJ. Populus: a model system for plant biology. Ann Rev of Plant Biol. 2007;58:3485–3501. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

