Modeling sample variables with an Experimental Factor Ontology
- PMID: 20200009
- PMCID: PMC2853691
- DOI: 10.1093/bioinformatics/btq099
Modeling sample variables with an Experimental Factor Ontology
Abstract
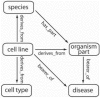
Motivation: Describing biological sample variables with ontologies is complex due to the cross-domain nature of experiments. Ontologies provide annotation solutions; however, for cross-domain investigations, multiple ontologies are needed to represent the data. These are subject to rapid change, are often not interoperable and present complexities that are a barrier to biological resource users.
Results: We present the Experimental Factor Ontology, designed to meet cross-domain, application focused use cases for gene expression data. We describe our methodology and open source tools used to create the ontology. These include tools for creating ontology mappings, ontology views, detecting ontology changes and using ontologies in interfaces to enhance querying. The application of reference ontologies to data is a key problem, and this work presents guidelines on how community ontologies can be presented in an application ontology in a data-driven way.
Availability: http://www.ebi.ac.uk/efo.
Figures




References
-
- Blake JA, Harris MA. The Gene Ontology (GO) project: structured vocabularies for molecular biology and their application to genome and expression analysis. Curr. Protoc. Bioinformatics. 2008 Chapter 7, Unit 7.2. - PubMed
-
- Bizer C, et al. Linked data - the story so far. Int. J. Semant. Web Inf. Syst. 2009;5:1–22.
-
- Grenon P, Smith B. SNAP and SPAN: towards dynamic spatial ontology. Spat. Cogn. Comput.Interdiscip. J. 2004;4:69–104.
-
- Gómez-Pérez A, et al. Ontological engineering. New York: Springer-Verlag; 2004.
-
- Horridge M. The OWL API: A Java API for Working with OWL 2 Ontologies. Proceedings of the OWL: Experiences and Directions 2009, Chantilly, USA, CEUR Workshop Proceedings. 2009
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

