Distinct RNA-dependent RNA polymerases are required for RNAi triggered by double-stranded RNA versus truncated transgenes in Paramecium tetraurelia
- PMID: 20200046
- PMCID: PMC2896523
- DOI: 10.1093/nar/gkq131
Distinct RNA-dependent RNA polymerases are required for RNAi triggered by double-stranded RNA versus truncated transgenes in Paramecium tetraurelia
Abstract
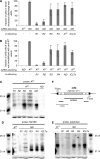
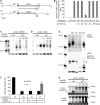
In many eukaryotes, RNA-dependent RNA polymerases (RdRPs) play key roles in the RNAi pathway. They have been implicated in the recognition and processing of aberrant transcripts triggering the process, and in amplification of the silencing response. We have tested the functions of RdRP genes from the ciliate Paramecium tetraurelia in experimentally induced and endogenous mechanisms of gene silencing. In this organism, RNAi can be triggered either by high-copy, truncated transgenes or by directly feeding cells with double-stranded RNA (dsRNA). Surprisingly, dsRNA-induced silencing depends on the putatively functional RDR1 and RDR2 genes, which are required for the accumulation of both primary siRNAs and a distinct class of small RNAs suggestive of secondary siRNAs. In contrast, a third gene with a highly divergent catalytic domain, RDR3, is required for siRNA accumulation when RNAi is triggered by truncated transgenes. Our data further implicate RDR3 in the accumulation of previously described endogenous siRNAs and in the regulation of the surface antigen gene family. While only one of these genes is normally expressed in any clonal cell line, the knockdown of RDR3 leads to co-expression of multiple antigens. These results provide evidence for a functional specialization of Paramecium RdRP genes in distinct RNAi pathways operating during vegetative growth.
Figures


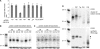

Similar articles
-
Genomic characterization of variable surface antigens reveals a telomere position effect as a prerequisite for RNA interference-mediated silencing in Paramecium tetraurelia.mBio. 2014 Nov 11;5(6):e01328. doi: 10.1128/mBio.01328-14. mBio. 2014. PMID: 25389173 Free PMC article.
-
A forward genetic screen reveals essential and non-essential RNAi factors in Paramecium tetraurelia.Nucleic Acids Res. 2014 Jun;42(11):7268-80. doi: 10.1093/nar/gku223. Epub 2014 May 23. Nucleic Acids Res. 2014. PMID: 24860163 Free PMC article.
-
Primary and secondary siRNA synthesis triggered by RNAs from food bacteria in the ciliate Paramecium tetraurelia.Nucleic Acids Res. 2015 Feb 18;43(3):1818-33. doi: 10.1093/nar/gku1331. Epub 2015 Jan 15. Nucleic Acids Res. 2015. PMID: 25593325 Free PMC article.
-
The long and short of siRNAs.Mol Cell. 2002 Sep;10(3):435-7. doi: 10.1016/s1097-2765(02)00657-3. Mol Cell. 2002. PMID: 12408811 Review.
-
RNA interference: the molecular immune system.J Mol Histol. 2004 Aug;35(6):545-53. doi: 10.1007/s10735-004-2192-8. J Mol Histol. 2004. PMID: 15614608 Review.
Cited by
-
Genomic characterization of variable surface antigens reveals a telomere position effect as a prerequisite for RNA interference-mediated silencing in Paramecium tetraurelia.mBio. 2014 Nov 11;5(6):e01328. doi: 10.1128/mBio.01328-14. mBio. 2014. PMID: 25389173 Free PMC article.
-
The 5' spreading of small RNAs in Dictyostelium discoideum depends on the RNA-dependent RNA polymerase RrpC and on the dicer-related nuclease DrnB.PLoS One. 2013 May 20;8(5):e64804. doi: 10.1371/journal.pone.0064804. Print 2013. PLoS One. 2013. PMID: 23724097 Free PMC article.
-
RNAi effector diversity in nematodes.PLoS Negl Trop Dis. 2011 Jun;5(6):e1176. doi: 10.1371/journal.pntd.0001176. Epub 2011 Jun 7. PLoS Negl Trop Dis. 2011. PMID: 21666793 Free PMC article.
-
Functional lability of RNA-dependent RNA polymerases in animals.PLoS Genet. 2019 Feb 19;15(2):e1007915. doi: 10.1371/journal.pgen.1007915. eCollection 2019 Feb. PLoS Genet. 2019. PMID: 30779744 Free PMC article.
-
Two sets of RNAi components are required for heterochromatin formation in trans triggered by truncated transgenes.Nucleic Acids Res. 2016 Jul 8;44(12):5908-23. doi: 10.1093/nar/gkw267. Epub 2016 Apr 16. Nucleic Acids Res. 2016. PMID: 27085807 Free PMC article.
References
-
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. - PubMed
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. - PubMed
-
- Chan SW, Zilberman D, Xie Z, Johansen LK, Carrington JC, Jacobsen SE. RNA silencing genes control de novo DNA methylation. Science. 2004;303:1336. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

