The regulation of MADS-box gene expression during ripening of banana and their regulatory interaction with ethylene
- PMID: 20200120
- PMCID: PMC2837265
- DOI: 10.1093/jxb/erq017
The regulation of MADS-box gene expression during ripening of banana and their regulatory interaction with ethylene
Abstract
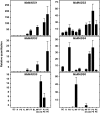
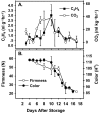
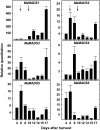
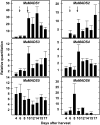
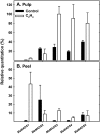
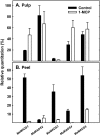
Six MaMADS-box genes have been cloned from the banana fruit cultivar Grand Nain. The similarity of these genes to tomato LeRIN is low and neither MaMADS2 nor MaMADS1 complement the tomato rin mutation. Nevertheless, the expression patterns, specifically in fruit and the induction during ripening and in response to ethylene and 1-MCP, suggest that some of these genes may participate in ripening. MaMADS1, 2, and 3, are highly expressed in fruit only, while the others are expressed in fruit as well as in other organs. Moreover, the suites of MaMADS-box genes and their temporal expression differ in peel and pulp during ripening. In the pulp, the increase in MaMADS2, 3, 4, and 5 expression preceded an increase in ethylene production, but coincides with the CO(2) peak. However, MaMADS1 expression in pulp coincided with ethylene production, but a massive increase in its expression occurred late during ripening, together with a second wave in the expression of MaMADS2, 3, and 4. In the peel, on the other hand, an increase in expression of MaMADS1, 3, and to a lesser degree also of MaMADS4 and 2 coincided with an increase in ethylene production. Except MaMADS3, which was induced by ethylene in pulp and peel, only MaMADS4, and 5 in pulp and MaMADS1 in peel were induced by ethylene. 1-MCP applied at the onset of the increase in ethylene production, increased the levels of MaMADS4 and MaMADS1 in pulp, while it decreased MaMADS1, 3, 4, and 5 in peel, suggesting that MaMADS4 and MaMADS1 are negatively controlled by ethylene at the onset of ethylene production only in pulp. Only MaMADS2 is neither induced by ethylene nor by 1-MCP, and it is expressed mainly in pulp. Our results suggest that two independent ripening programs are employed in pulp and peel which involve the activation of mainly MaMADS2, 4, and 5 and later on also MaMADS1 in pulp, and mainly MaMADS1, and 3 in peel. Hence, our results are consistent with MaMADS2, a SEP3 homologue, acting in the pulp upstream of the increase in ethylene production similarly to LeMADS-RIN.
Figures


Similar articles
-
Banana MaMADS Transcription Factors Are Necessary for Fruit Ripening and Molecular Tools to Promote Shelf-Life and Food Security.Plant Physiol. 2016 May;171(1):380-91. doi: 10.1104/pp.15.01866. Epub 2016 Mar 8. Plant Physiol. 2016. PMID: 26956665 Free PMC article.
-
The MaNAP1-MaMADS1 transcription factor module mediates ethylene-regulated peel softening and ripening in banana.Plant Cell. 2024 Dec 23;37(1):koae282. doi: 10.1093/plcell/koae282. Plant Cell. 2024. PMID: 39422253
-
Differential feedback regulation of ethylene biosynthesis in pulp and peel tissues of banana fruit.J Exp Bot. 2007;58(5):1047-57. doi: 10.1093/jxb/erl265. Epub 2006 Dec 21. J Exp Bot. 2007. PMID: 17185740
-
Variations on a theme in fruit development: the PLE lineage of MADS-box genes in tomato (TAGL1) and other species.Planta. 2017 Aug;246(2):313-321. doi: 10.1007/s00425-017-2725-5. Epub 2017 Jun 28. Planta. 2017. PMID: 28660293 Review.
-
Use of genomics tools to isolate key ripening genes and analyse fruit maturation in tomato.J Exp Bot. 2002 Oct;53(377):2023-30. doi: 10.1093/jxb/erf057. J Exp Bot. 2002. PMID: 12324526 Review.
Cited by
-
The ambiguous ripening nature of the fig (Ficus carica L.) fruit: a gene-expression study of potential ripening regulators and ethylene-related genes.J Exp Bot. 2015 Jun;66(11):3309-24. doi: 10.1093/jxb/erv140. Epub 2015 May 8. J Exp Bot. 2015. PMID: 25956879 Free PMC article.
-
Comparative transcriptional profiling analysis of developing melon (Cucumis melo L.) fruit from climacteric and non-climacteric varieties.BMC Genomics. 2015 Jun 9;16(1):440. doi: 10.1186/s12864-015-1649-3. BMC Genomics. 2015. PMID: 26054931 Free PMC article.
-
Characterizing the involvement of FaMADS9 in the regulation of strawberry fruit receptacle development.Plant Biotechnol J. 2020 Apr;18(4):929-943. doi: 10.1111/pbi.13257. Epub 2019 Oct 11. Plant Biotechnol J. 2020. PMID: 31533196 Free PMC article.
-
MaMADS2 repression in banana fruits modifies hormone synthesis and signalling pathways prior to climacteric stage.BMC Plant Biol. 2018 Nov 6;18(1):267. doi: 10.1186/s12870-018-1480-5. BMC Plant Biol. 2018. PMID: 30400866 Free PMC article.
-
Flower development of Phalaenopsis orchid involves functionally divergent SEPALLATA-like genes.New Phytol. 2014 May;202(3):1024-1042. doi: 10.1111/nph.12723. Epub 2014 Feb 14. New Phytol. 2014. PMID: 24571782 Free PMC article.
References
-
- Adam H, Jouannic S, Morcillo F, Richaud F, Duval Y, Tregear J. MADS box genes in oil palm (Elaeis guineensis): patterns in the evolution of the SQUAMOSA, DEFICIENS, GLOBOSA, AGAMOUS, and SEPALLATA subfamilies. Journal of Molecular Evolution. 2006;62:15–31. - PubMed
-
- Adams-Phillips L, Barry C, Giovannoni J. Signal transduction systems regulating fruit ripening. Trends in Plant Science. 2004;9:331–338. - PubMed
-
- Alexander L, Grierson D. Ethylene biosynthesis and action in tomato:a model for climacteric fruit ripening. Journal of Experimental Botany. 2002;53:2039–2055. - PubMed
-
- Andersen CH, Jensen CS, Petersen K. Similar genetic switch systems might integrate the floral inductive pathways in dicots and monocots. Trends in Plant Science. 2004;9:105–107. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

