Gene expression profiling indicates the roles of host oxidative stress, apoptosis, lipid metabolism, and intracellular transport genes in the replication of hepatitis C virus
- PMID: 20200238
- PMCID: PMC2863852
- DOI: 10.1128/JVI.02529-09
Gene expression profiling indicates the roles of host oxidative stress, apoptosis, lipid metabolism, and intracellular transport genes in the replication of hepatitis C virus
Abstract
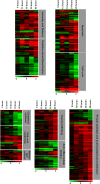
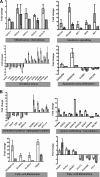
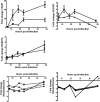
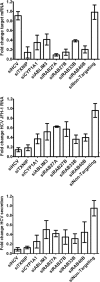
Hepatitis C virus (HCV) is a leading cause of chronic liver disease. The identification and characterization of key host cellular factors that play a role in the HCV replication cycle are important for the understanding of disease pathogenesis and the identification of novel antiviral therapeutic targets. Gene expression profiling of JFH-1-infected Huh7 cells by microarray analysis was performed to identify host cellular genes that are transcriptionally regulated by infection. The expression of host genes involved in cellular defense mechanisms (apoptosis, proliferation, and antioxidant responses), cellular metabolism (lipid and protein metabolism), and intracellular transport (vesicle trafficking and cytoskeleton regulation) was significantly altered by HCV infection. The gene expression patterns identified provide insight into the potential mechanisms that contribute to HCV-associated pathogenesis. These include an increase in proinflammatory and proapoptotic signaling and a decrease in the antioxidant response pathways of the infected cell. To investigate whether any of the host genes regulated by infection were required by HCV during replication, small interfering RNA (siRNA) silencing of host gene expression in HCV-infected cells was performed. Decreasing the expression of host genes involved in lipid metabolism (TXNIP and CYP1A1 genes) and intracellular transport (RAB33b and ABLIM3 genes) reduced the replication and secretion of HCV, indicating that they may be important factors for the virus replication cycle. These results show that major changes in the expression of many different genes in target cells may be crucial in determining the outcome of HCV infection.
Figures
Similar articles
-
N-Myc Downstream-Regulated Gene 1 Restricts Hepatitis C Virus Propagation by Regulating Lipid Droplet Biogenesis and Viral Assembly.J Virol. 2018 Jan 2;92(2):e01166-17. doi: 10.1128/JVI.01166-17. Print 2018 Jan 15. J Virol. 2018. PMID: 29118118 Free PMC article.
-
A host cell RNA-binding protein, Staufen1, has a role in hepatitis C virus replication before virus assembly.J Gen Virol. 2013 Nov;94(Pt 11):2429-2436. doi: 10.1099/vir.0.051383-0. Epub 2013 Aug 1. J Gen Virol. 2013. PMID: 23907398
-
[Inhibition of silent information regulator-1 in hepatocytes induces lipid metabolism disorders and enhances hepatitis C virus replication].Zhonghua Gan Zang Bing Za Zhi. 2013 Nov;21(11):834-9. doi: 10.3760/cma.j.issn.1007-3418.2013.11.009. Zhonghua Gan Zang Bing Za Zhi. 2013. PMID: 24331693 Chinese.
-
Cellular stress responses in hepatitis C virus infection: Mastering a two-edged sword.Virus Res. 2015 Nov 2;209:100-17. doi: 10.1016/j.virusres.2015.03.013. Epub 2015 Mar 30. Virus Res. 2015. PMID: 25836277 Review.
-
Hepatitis C Virus Infection Induces Autophagy as a Prosurvival Mechanism to Alleviate Hepatic ER-Stress Response.Viruses. 2016 May 23;8(5):150. doi: 10.3390/v8050150. Viruses. 2016. PMID: 27223299 Free PMC article. Review.
Cited by
-
Transcriptome and miRNome Analysis Provide New Insight Into Host Lipid Accumulation, Innate Immunity, and Viral Persistence in Hepatitis C Virus Infection in vitro.Front Microbiol. 2020 Sep 30;11:535673. doi: 10.3389/fmicb.2020.535673. eCollection 2020. Front Microbiol. 2020. PMID: 33101221 Free PMC article.
-
Hepatitis B Virus Subverts the Autophagy Elongation Complex Atg5-12/16L1 and Does Not Require Atg8/LC3 Lipidation for Viral Maturation.J Virol. 2018 Mar 14;92(7):e01513-17. doi: 10.1128/JVI.01513-17. Print 2018 Apr 1. J Virol. 2018. PMID: 29367244 Free PMC article.
-
Cyclosporine A Suppressed Glucose Oxidase Induced P53 Mitochondrial Translocation and Hepatic Cell Apoptosis through Blocking Mitochondrial Permeability Transition.Int J Biol Sci. 2016 Jan 1;12(2):198-209. doi: 10.7150/ijbs.13716. eCollection 2016. Int J Biol Sci. 2016. PMID: 26884717 Free PMC article.
-
Regulation of hepatitis C virus replication by nuclear translocation of nonstructural 5A protein and transcriptional activation of host genes.J Virol. 2013 May;87(10):5523-39. doi: 10.1128/JVI.00585-12. Epub 2013 Mar 6. J Virol. 2013. PMID: 23468497 Free PMC article.
-
Identification and comparative analysis of hepatitis C virus-host cell protein interactions.Mol Biosyst. 2013 Dec;9(12):3199-209. doi: 10.1039/c3mb70343f. Epub 2013 Oct 18. Mol Biosyst. 2013. PMID: 24136289 Free PMC article.
References
-
- Abe, K., M. Ikeda, H. Dansako, K. Naka, K. Shimotohno, and N. Kato. 2005. cDNA microarray analysis to compare HCV subgenomic replicon cells with their cured cells. Virus Res. 107:73-81. - PubMed
-
- Aizaki, H., T. Harada, M. Otsuka, N. Seki, M. Matsuda, Y. W. Li, H. Kawakami, Y. Matsuura, T. Miyamura, and T. Suzuki. 2002. Expression profiling of liver cell lines expressing entire or parts of hepatitis C virus open reading frame. Hepatology 36:1431-1438. - PubMed
-
- Bartenschlager, R., and S. Sparacio. 2007. Hepatitis C virus molecular clones and their replication capacity in vivo and in cell culture. Virus Res. 127:195-207. - PubMed
-
- Basu, A., K. Meyer, K. K. Lai, K. Saito, A. M. Di Bisceglie, L. E. Grosso, R. B. Ray, and R. Ray. 2006. Microarray analyses and molecular profiling of Stat3 signaling pathway induced by hepatitis C virus core protein in human hepatocytes. Virology 349:347-358. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

