The de novo methyltransferases DNMT3a and DNMT3b target the murine gammaherpesvirus immediate-early gene 50 promoter during establishment of latency
- PMID: 20200245
- PMCID: PMC2863815
- DOI: 10.1128/JVI.00060-10
The de novo methyltransferases DNMT3a and DNMT3b target the murine gammaherpesvirus immediate-early gene 50 promoter during establishment of latency
Abstract
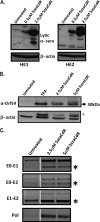
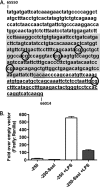
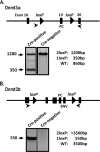
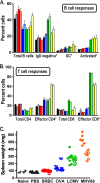
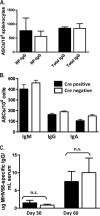
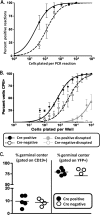
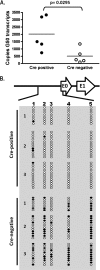
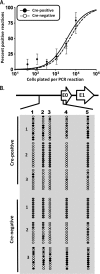
The role of epigenetic modifications in the regulation of gammaherpesvirus latency has been a subject of active study for more than 20 years. DNA methylation, associated with transcriptional silencing in mammalian genomes, has been shown to be an important mechanism in the transcriptional control of several key gammaherpesvirus genes. In particular, DNA methylation of the functionally conserved immediate-early replication and transcription activator (RTA) has been shown to regulate Epstein-Barr virus and Kaposi's sarcoma-associated herpesvirus Rta expression. Here we demonstrate that the murine gammaherpesvirus (MHV68) homolog, encoded by gene 50, is also subject to direct repression by DNA methylation, both in vitro and in vivo. We observed that the treatment of latently MHV68-infected B-cell lines with a methyltransferase inhibitor induced virus reactivation. In addition, we show that the methylation of the recently characterized distal gene 50 promoter represses activity in a murine macrophage cell line. To evaluate the role of de novo methyltransferases (DNMTs) in the establishment of these methylation marks, we infected mice in which conditional DNMT3a and DNMT3b alleles were selectively deleted in B lymphocytes. DNMT3a/DNMT3b-deficient B cells were phenotypically normal, displaying no obvious compromise in cell surface marker expression or antibody production either in naïve mice or in the context of nonviral and viral immunogens. However, mice lacking functional DNMT3a and DNMT3b in B cells exhibited hallmarks of deregulated MHV68 lytic replication, including increased splenomegaly and the presence of infectious virus in the spleen at day 18 following infection. In addition, total gene 50 transcript levels were elevated in the spleens of these mice at day 18, which correlated with the hypomethylation of the distal gene 50 promoter. However, by day 42 postinfection, aberrant virus replication was resolved, and we observed wild-type frequencies of viral genome-positive splenocytes in mice lacking functional DNMT3a and DNMT3b in B lymphocytes. The latter correlated with increased CpG methylation in the distal gene 50 promoter, which was restored to levels similar to those of littermate controls harboring functional DNMT3a and DNMT3b alleles in B lymphocytes, suggesting the existence of an alternative mechanism for the de novo methylation of the MHV68 genome. Importantly, this DNMT3a/DNMT3b-independent methylation appeared to be targeted specifically to the gene 50 promoter, as we observed that the promoters for MHV68 gene 72 (v-cyclin) and M11 (v-bcl2) remained hypomethylated at day 42 postinfection. Taken together, these data provide the first evidence of the importance of DNA methylation in regulating gammaherpesvirus RTA/gene 50 transcription during virus infection in vivo and provide insight into the hierarchy of host machinery required to establish this modification.
Figures

Similar articles
-
Alternatively initiated gene 50/RTA transcripts expressed during murine and human gammaherpesvirus reactivation from latency.J Virol. 2009 Jan;83(1):314-28. doi: 10.1128/JVI.01444-08. Epub 2008 Oct 29. J Virol. 2009. PMID: 18971285 Free PMC article.
-
Murine gammaherpesvirus 68 reactivation from B cells requires IRF4 but not XBP-1.J Virol. 2014 Oct;88(19):11600-10. doi: 10.1128/JVI.01876-14. Epub 2014 Jul 30. J Virol. 2014. PMID: 25078688 Free PMC article.
-
Lytic Replication and Reactivation from B Cells Is Not Required for Establishing or Maintaining Gammaherpesvirus Latency In Vivo.J Virol. 2022 Jun 22;96(12):e0069022. doi: 10.1128/jvi.00690-22. Epub 2022 Jun 1. J Virol. 2022. PMID: 35647668 Free PMC article.
-
Gamma interferon blocks gammaherpesvirus reactivation from latency in a cell type-specific manner.J Virol. 2007 Jun;81(11):6134-40. doi: 10.1128/JVI.00108-07. Epub 2007 Mar 14. J Virol. 2007. PMID: 17360749 Free PMC article. Review.
-
Murine Gammaherpesvirus 68: A Small Animal Model for Gammaherpesvirus-Associated Diseases.Adv Exp Med Biol. 2017;1018:225-236. doi: 10.1007/978-981-10-5765-6_14. Adv Exp Med Biol. 2017. PMID: 29052141 Review.
Cited by
-
The Interplay between Chromatin and Transcription Factor Networks during B Cell Development: Who Pulls the Trigger First?Front Immunol. 2014 Apr 11;5:156. doi: 10.3389/fimmu.2014.00156. eCollection 2014. Front Immunol. 2014. PMID: 24782862 Free PMC article. Review.
-
An in vitro system for studying murid herpesvirus-4 latency and reactivation.PLoS One. 2010 Jun 11;5(6):e11080. doi: 10.1371/journal.pone.0011080. PLoS One. 2010. PMID: 20552028 Free PMC article.
-
Pervasive transcription of a herpesvirus genome generates functionally important RNAs.mBio. 2014 Mar 11;5(2):e01033-13. doi: 10.1128/mBio.01033-13. mBio. 2014. PMID: 24618256 Free PMC article.
-
Interferon regulatory factor 8 regulates caspase-1 expression to facilitate Epstein-Barr virus reactivation in response to B cell receptor stimulation and chemical induction.PLoS Pathog. 2018 Jan 22;14(1):e1006868. doi: 10.1371/journal.ppat.1006868. eCollection 2018 Jan. PLoS Pathog. 2018. PMID: 29357389 Free PMC article.
-
KSHV LANA--the master regulator of KSHV latency.Viruses. 2014 Dec 11;6(12):4961-98. doi: 10.3390/v6124961. Viruses. 2014. PMID: 25514370 Free PMC article. Review.
References
-
- Barozzi, P., L. Potenza, G. Riva, D. Vallerini, C. Quadrelli, R. Bosco, F. Forghieri, G. Torelli, and M. Luppi. 2007. B cells and herpesviruses: a model of lymphoproliferation. Autoimmun. Rev. 7:132-136. - PubMed
-
- Ben-Sasson, S. A., and G. Klein. 1981. Activation of the Epstein-Barr virus genome by 5-aza-cytidine in latently infected human lymphoid lines. Int. J. Cancer 28:131-135. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

