The first complete papillomavirus genome characterized from a marsupial host: a novel isolate from Bettongia penicillata
- PMID: 20200246
- PMCID: PMC2863809
- DOI: 10.1128/JVI.02635-09
The first complete papillomavirus genome characterized from a marsupial host: a novel isolate from Bettongia penicillata
Abstract
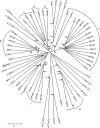
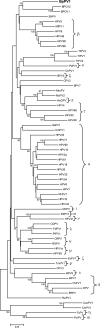
The first fully sequenced papillomavirus (PV) of marsupials, tentatively named Bettongia penicillata papillomavirus type 1 (BpPV1), was detected in papillomas from a woylie (Bettongia penicillata ogilbyi). The circular, double-stranded DNA genome contains 7,737 bp and encodes 7 open reading frames (ORFs), E6, E7, E1, E2, E4, L2, and L1, in typical PV conformation. BpPV1 is a close-to-root PV with L1 and L2 ORFs most similar to European hedgehog PV and bandicoot papillomatosis carcinomatosis virus types 1 and 2 (BPCV1 and -2). It appears that the BPCVs arose by recombination between an ancient PV and an ancient polyomavirus more than 10 million years ago.
Figures

Similar articles
-
Genomic characterization of a novel virus found in papillomatous lesions from a southern brown bandicoot (Isoodon obesulus) in Western Australia.Virology. 2008 Jun 20;376(1):173-82. doi: 10.1016/j.virol.2008.03.014. Epub 2008 Apr 25. Virology. 2008. PMID: 18440042
-
Whole-genome characterization of a novel polyomavirus detected in fatally diseased canary birds.J Gen Virol. 2010 Dec;91(Pt 12):3016-22. doi: 10.1099/vir.0.023549-0. Epub 2010 Aug 25. J Gen Virol. 2010. PMID: 20797969
-
Discovery of a polyomavirus in European badgers (Meles meles) and the evolution of host range in the family Polyomaviridae.J Gen Virol. 2015 Jun;96(Pt 6):1411-1422. doi: 10.1099/vir.0.000071. Epub 2015 Jan 27. J Gen Virol. 2015. PMID: 25626684 Free PMC article.
-
Genome analysis of the new human polyomaviruses.Rev Med Virol. 2012 Nov;22(6):354-77. doi: 10.1002/rmv.1711. Epub 2012 Mar 28. Rev Med Virol. 2012. PMID: 22461085 Review.
-
Genome analysis of non-human primate polyomaviruses.Infect Genet Evol. 2014 Aug;26:283-94. doi: 10.1016/j.meegid.2014.05.030. Epub 2014 Jun 14. Infect Genet Evol. 2014. PMID: 24933462 Review.
Cited by
-
Identification of a novel polyomavirus from a marsupial host.Virus Evol. 2022 Oct 6;8(2):veac096. doi: 10.1093/ve/veac096. eCollection 2022. Virus Evol. 2022. PMID: 36381233 Free PMC article.
-
Temporal Patterns in the Abundance of a Critically Endangered Marsupial Relates to Disturbance by Roads and Agriculture.PLoS One. 2016 Aug 8;11(8):e0160790. doi: 10.1371/journal.pone.0160790. eCollection 2016. PLoS One. 2016. PMID: 27501320 Free PMC article.
-
Widespread Horizontal Gene Transfer Among Animal Viruses.bioRxiv [Preprint]. 2024 Mar 26:2024.03.25.586562. doi: 10.1101/2024.03.25.586562. bioRxiv. 2024. PMID: 38712252 Free PMC article. Preprint.
-
Classification and nomenclature system for human Alphapapillomavirus variants: general features, nucleotide landmarks and assignment of HPV6 and HPV11 isolates to variant lineages.Acta Dermatovenerol Alp Pannonica Adriat. 2011 Sep;20(3):113-23. Acta Dermatovenerol Alp Pannonica Adriat. 2011. PMID: 22131111 Free PMC article.
-
Association of papillomavirus E6 proteins with either MAML1 or E6AP clusters E6 proteins by structure, function, and evolutionary relatedness.PLoS Pathog. 2017 Dec 27;13(12):e1006781. doi: 10.1371/journal.ppat.1006781. eCollection 2017 Dec. PLoS Pathog. 2017. PMID: 29281732 Free PMC article.
References
-
- Antonsson, A., and A. J. McMillan. 2006. Papillomavirus in healthy skin of Australian animals. J. Gen. Virol. 87:3195-3200. - PubMed
-
- Bennett, M. D., L. Woolford, A. J. O'Hara, K. S. Warren, and P. K. Nicholls. 2008. In situ hybridization to detect bandicoot papillomatosis carcinomatosis virus type 1 in biopsies from endangered western barred bandicoots (Perameles bougainville). J. Gen. Virol. 89:419-423. - PubMed
-
- Bennett, M. D., L. Woolford, H. Stevens, M. Van Ranst, T. Oldfield, M. Slaven, A. J. O'Hara, K. S. Warren, and P. K. Nicholls. 2008. Genomic characterization of a novel virus found in papillomatous lesions from a southern brown bandicoot (Isoodon obesulus) in Western Australia. Virology 376:173-182. - PubMed
-
- Bernard, H.-U. 2008. Genome diversity and evolution of papillomaviruses, p. 417-429. In E. Domingo, C. Parish, and J. Holland (ed.), Origin and evolution of viruses, 2nd ed. Academic Press, London, England.
-
- Christensen, P. 2002. Brush-tailed bettong, p. 292-293. In R. Strahan (ed.), The mammals of Australia, 2nd ed. Reed New Holland, Frenchs Forest, New South Wales, Australia.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

