Regulation of Epstein-Barr virus origin of plasmid replication (OriP) by the S-phase checkpoint kinase Chk2
- PMID: 20200249
- PMCID: PMC2863808
- DOI: 10.1128/JVI.01300-09
Regulation of Epstein-Barr virus origin of plasmid replication (OriP) by the S-phase checkpoint kinase Chk2
Abstract
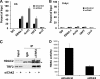
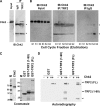
The Epstein-Barr virus (EBV) origin of plasmid replication (OriP) is required for episome stability during latent infection. Telomere repeat factor 2 (TRF2) binds directly to OriP and facilitates DNA replication and plasmid maintenance. Recent studies have found that TRF2 interacts with the DNA damage checkpoint protein Chk2. We show here that Chk2 plays an important role in regulating OriP plasmid stability, chromatin modifications, and replication timing. The depletion of Chk2 by small interfering RNA (siRNA) leads to a reduction in DNA replication efficiency and a loss of OriP-dependent plasmid maintenance. This corresponds to a change in OriP replication timing and an increase in constitutive histone H3 acetylation. We show that Chk2 interacts with TRF2 in the early G(1)/S phase of the cell cycle. We also show that Chk2 can phosphorylate TRF2 in vitro at a consensus acceptor site in the amino-terminal basic domain of TRF2. TRF2 mutants with a serine-to-aspartic acid phosphomimetic substitution mutation were reduced in their ability to recruit the origin recognition complex (ORC) and stimulate OriP replication. We suggest that the Chk2 phosphorylation of TRF2 is important for coordinating ORC binding with chromatin remodeling during the early S phase and that a failure to execute these events leads to replication defects and plasmid instability.
Figures





Similar articles
-
Regulation of Epstein-Barr virus OriP replication by poly(ADP-ribose) polymerase 1.J Virol. 2010 May;84(10):4988-97. doi: 10.1128/JVI.02333-09. Epub 2010 Mar 10. J Virol. 2010. PMID: 20219917 Free PMC article.
-
Telomere repeat binding factors TRF1, TRF2, and hRAP1 modulate replication of Epstein-Barr virus OriP.J Virol. 2003 Nov;77(22):11992-2001. doi: 10.1128/jvi.77.22.11992-12001.2003. J Virol. 2003. PMID: 14581536 Free PMC article.
-
Epstein-Barr virus episome stability is coupled to a delay in replication timing.J Virol. 2009 Mar;83(5):2154-62. doi: 10.1128/JVI.02115-08. Epub 2008 Dec 10. J Virol. 2009. PMID: 19073720 Free PMC article.
-
EBNA1 and host factors in Epstein-Barr virus latent DNA replication.Curr Opin Virol. 2012 Dec;2(6):733-9. doi: 10.1016/j.coviro.2012.09.005. Epub 2012 Sep 30. Curr Opin Virol. 2012. PMID: 23031715 Review.
-
Latent and lytic Epstein-Barr virus replication strategies.Rev Med Virol. 2005 Jan-Feb;15(1):3-15. doi: 10.1002/rmv.441. Rev Med Virol. 2005. PMID: 15386591 Review.
Cited by
-
Epstein-Barr nuclear antigen 1 (EBNA1)-dependent recruitment of origin recognition complex (Orc) on oriP of Epstein-Barr virus with purified proteins: stimulation by Cdc6 through its direct interaction with EBNA1.J Biol Chem. 2012 Jul 6;287(28):23977-94. doi: 10.1074/jbc.M112.368456. Epub 2012 May 14. J Biol Chem. 2012. PMID: 22589552 Free PMC article.
-
The origin recognition complex in human diseases.Biosci Rep. 2013 Jun 11;33(3):e00044. doi: 10.1042/BSR20130036. Biosci Rep. 2013. PMID: 23662735 Free PMC article. Review.
-
Interplay between DNA tumor viruses and the host DNA damage response.Curr Top Microbiol Immunol. 2013;371:229-57. doi: 10.1007/978-3-642-37765-5_9. Curr Top Microbiol Immunol. 2013. PMID: 23686238 Free PMC article. Review.
-
TRF1 phosphorylation on T271 modulates telomerase-dependent telomere length maintenance as well as the formation of ALT-associated PML bodies.Sci Rep. 2016 Nov 14;6:36913. doi: 10.1038/srep36913. Sci Rep. 2016. PMID: 27841304 Free PMC article.
-
The Epstein-Barr Virus EBNA1 Protein.Scientifica (Cairo). 2012;2012:438204. doi: 10.6064/2012/438204. Epub 2012 Dec 19. Scientifica (Cairo). 2012. PMID: 24278697 Free PMC article. Review.
References
-
- Bell, S. P. 2002. The origin recognition complex: from simple origins to complex functions. Genes Dev. 16:659-672. - PubMed
-
- Buscemi, G., L. Zannini, E. Fontanella, D. Lecis, S. Lisanti, and D. Delia. 2009. The shelterin protein TRF2 inhibits Chk2 activity at telomeres in the absence of DNA damage. Curr. Biol. 19:874-879. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

