PARP inhibition: PARP1 and beyond
- PMID: 20200537
- PMCID: PMC2910902
- DOI: 10.1038/nrc2812
PARP inhibition: PARP1 and beyond
Abstract
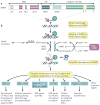
Recent findings have thrust poly(ADP-ribose) polymerases (PARPs) into the limelight as potential chemotherapeutic targets. To provide a framework for understanding these recent observations, we review what is known about the structures and functions of the family of PARP enzymes, and then outline a series of questions that should be addressed to guide the rational development of PARP inhibitors as anticancer agents.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
References
-
- Chambon P, Weill JD, Mandel P. Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem Biophys Res Commun. 1963;11:39–43. - PubMed
-
- Doly J, Petek F. Etude de la structure d’un composé “poly(ADP-ribose)” synthétisé par des extraits nucléaires de foie de poulet. C R Hebd Seances Acad Sci Ser D Sci Nat. 1966;263:1341–1344.
-
- Chambon P, Weill JD, Doly J, Strosser MT, Mandel P. On the formation of a novel adelylic compound by enzymatic extracts of liver nuclei. Biochem Biophys Res Commun. 1966;25:638–643.
-
- Nishizuka Y, Ueda K, Nakazawa K, Hayaishi O. Studies on the polymer of adenosine diphosphate ribose. I. Enzymic formation from nicotinamide adenine dinuclotide in mammalian nuclei. J Biol Chem. 1967;242:3164–3171. - PubMed
-
- Sugimura T, Fujimura S, Hasegawa S, Kawamura Y. Polymerization of the adenosine 5′-diphosphate ribose moiety of NAD by rat liver nuclear enzyme. Biochim Biophys Acta. 1967;138:438–441. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

