Identification of a distant cis-regulatory element controlling pharyngeal arch-specific expression of zebrafish gdf6a/radar
- PMID: 20201106
- PMCID: PMC3110066
- DOI: 10.1002/dvdy.22251
Identification of a distant cis-regulatory element controlling pharyngeal arch-specific expression of zebrafish gdf6a/radar
Abstract
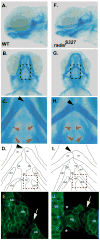
Skeletal formation is an essential and intricately regulated part of vertebrate development. Humans and mice deficient in growth and differentiation factor 6 (Gdf6) have numerous skeletal abnormalities, including joint fusions and cartilage reductions. The expression of Gdf6 is dynamic and in part regulated by distant evolutionarily conserved cis-regulatory elements. radar/gdf6a is a zebrafish ortholog of Gdf6 and has an essential role in embryonic patterning. Here, we show that radar is transcribed in the cells surrounding and between the developing cartilages of the ventral pharyngeal arches, similar to mouse Gdf6. A 312 bp evolutionarily conserved region (ECR5), 122 kilobases downstream, drives expression in a pharyngeal arch-specific manner similar to endogenous radar/gdf6a. Deletion analysis identified a 78 bp region within ECR5 that is essential for transgene activity. This work illustrates that radar is regulated in the pharyngeal arches by a distant conserved element and suggests radar has similar functions in skeletal development in fish and mammals.
Figures





Similar articles
-
Identification and characterization of the zebrafish pharyngeal arch-specific enhancer for the basic helix-loop-helix transcription factor Hand2.Dev Biol. 2012 Aug 1;368(1):118-26. doi: 10.1016/j.ydbio.2012.05.003. Epub 2012 May 14. Dev Biol. 2012. PMID: 22595513 Free PMC article.
-
Foxi transcription factors promote pharyngeal arch development by regulating formation of FGF signaling centers.Dev Biol. 2014 Jun 1;390(1):1-13. doi: 10.1016/j.ydbio.2014.03.004. Epub 2014 Mar 18. Dev Biol. 2014. PMID: 24650709 Free PMC article.
-
The lineage-specific gene ponzr1 is essential for zebrafish pronephric and pharyngeal arch development.Development. 2012 Feb;139(4):793-804. doi: 10.1242/dev.071720. Development. 2012. PMID: 22274699 Free PMC article.
-
Pathways in blood and vessel development revealed through zebrafish genetics.Int J Dev Biol. 2002;46(4):493-502. Int J Dev Biol. 2002. PMID: 12141436 Review.
-
Different words, same meaning: understanding the languages of the genome.Trends Genet. 2006 Dec;22(12):639-41. doi: 10.1016/j.tig.2006.09.009. Epub 2006 Sep 28. Trends Genet. 2006. PMID: 17010471 Review.
Cited by
-
Regulation of zebrafish melanocyte development by ligand-dependent BMP signaling.Elife. 2019 Dec 23;8:e50047. doi: 10.7554/eLife.50047. Elife. 2019. PMID: 31868592 Free PMC article.
-
The master male sex determinant Gdf6Y of the turquoise killifish arose through allelic neofunctionalization.Nat Commun. 2025 Jan 9;16(1):540. doi: 10.1038/s41467-025-55899-7. Nat Commun. 2025. PMID: 39788971 Free PMC article.
-
Stratified whole genome linkage analysis of Chiari type I malformation implicates known Klippel-Feil syndrome genes as putative disease candidates.PLoS One. 2013 Apr 19;8(4):e61521. doi: 10.1371/journal.pone.0061521. Print 2013. PLoS One. 2013. PMID: 23620759 Free PMC article.
-
Building and maintaining joints by exquisite local control of cell fate.Wiley Interdiscip Rev Dev Biol. 2017 Jan;6(1):10.1002/wdev.245. doi: 10.1002/wdev.245. Epub 2016 Sep 1. Wiley Interdiscip Rev Dev Biol. 2017. PMID: 27581688 Free PMC article. Review.
-
Transcriptional Interference Regulates the Evolutionary Development of Speech.Genes (Basel). 2022 Jul 4;13(7):1195. doi: 10.3390/genes13071195. Genes (Basel). 2022. PMID: 35885978 Free PMC article.
References
-
- Angelo S, Lohr J, Lee KH, Ticho BS, Breitbart RE, Hill S, Yost HJ, Srivastava D. Conservation of sequence and expression of Xenopus and zebrafish dHAND during cardiac, branchial arch and lateral mesoderm development. Mech Dev. 2000;95:231–237. - PubMed
-
- Asai-Coakwell M, French CR, Ye M, Garcha K, Bigot K, Perera AG, Staehling-Hampton K, Mema SC, Chanda B, Mushegian A, Bamforth S, Doschak MR, Li G, Dobbs MB, Giampietro PF, Brooks BP, Vijayalakshmi P, Sauve Y, Abitbol M, Sundaresan P, Heyningen VV, Pourquie O, Underhill TM, Waskiewicz AJ, Lehmann OJ. Incomplete penetrance and phenotypic variability characterize Gdf6-attributable oculo-skeletal phenotypes. Hum Mol Genet. 2009;18:1110–1121. - PubMed
-
- Bruneau S, Mourrain P, Rosa FM. Expression of contact, a new zebrafish DVR member, marks mesenchymal cell lineages in the developing pectoral fins and head and is regulated by retinoic acid. Mech Dev. 1997;65:163–173. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

