Nanoparticle detection of respiratory infection
- PMID: 20201109
- PMCID: PMC7169802
- DOI: 10.1002/wnan.83
Nanoparticle detection of respiratory infection
Abstract
Respiratory viruses are a constant concern for all demographics. Examples include established viruses such as respiratory syncytial virus (RSV), the leading cause of respiratory infection in infants and young children, and emerging viruses such as severe acute respiratory syndrome (SARS), which reached near pandemic levels in 2003, or H1N1 (swine) influenza. Despite this prevalence, traditional methods of virus detection are typically labor intensive and require several days to successfully confirm infection. Recently, however, nanoparticle-based detection strategies have been employed in an effort to develop detection assays that are both sensitive and expedient. Each of these platforms capitalizes on the unique properties of nanoparticles for the detection of respiratory viruses. In this article, several nanoparticle-based scaffolds are discussed. Gold nanoparticles (AuNPs) have been functionalized with virus specific antibodies or oligonucleotides. In each of these constructs, AuNPs act as both an easily conjugated scaffolding system for biological molecules and a powerful fluorescence quencher. AuNPs have also been immobilized and used as electrochemical transducers. They efficiently serve as a conducting interface of electrocatalyic activity making them a powerful tool in this application. Quantum dots (QDs) posses unique fluorescence properties that have also been explored for their application to virus detection when combined with direct antibody conjugation or streptavidin-biotin binding systems. QDs have an advantage over many traditional fluorophores because their fluorescence properties can be finely tuned and they are resistant to photobleaching. The development of these nanoparticle-based detection strategies holds the potential to be a powerful method to quickly and easily confirm respiratory virus infection.
Figures

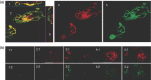
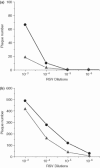
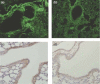
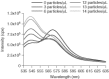

Similar articles
-
Molecular diagnosis of respiratory virus infections.Crit Rev Clin Lab Sci. 2011 Sep-Dec;48(5-6):217-49. doi: 10.3109/10408363.2011.640976. Crit Rev Clin Lab Sci. 2011. PMID: 22185616 Review.
-
Versatility of a localized surface plasmon resonance-based gold nanoparticle-alloyed quantum dot nanobiosensor for immunofluorescence detection of viruses.Biosens Bioelectron. 2017 Mar 15;89(Pt 2):998-1005. doi: 10.1016/j.bios.2016.10.045. Epub 2016 Oct 20. Biosens Bioelectron. 2017. PMID: 27825520
-
Molecular detection of respiratory viruses in clinical specimens from children with acute respiratory disease in Iran.Pediatr Infect Dis J. 2010 Oct;29(10):931-3. doi: 10.1097/inf.0b013e3181e2062e. Pediatr Infect Dis J. 2010. PMID: 20879092
-
Optimized detection of respiratory viruses in nasopharyngeal secretions.New Microbiol. 2003 Apr;26(2):133-40. New Microbiol. 2003. PMID: 12737194
-
Nucleic acid amplification-based diagnosis of respiratory virus infections.Expert Rev Anti Infect Ther. 2010 Nov;8(11):1273-92. doi: 10.1586/eri.10.121. Expert Rev Anti Infect Ther. 2010. PMID: 21073292 Review.
Cited by
-
Electrochemical diagnostics of infectious viral diseases: Trends and challenges.Biosens Bioelectron. 2021 May 15;180:113112. doi: 10.1016/j.bios.2021.113112. Epub 2021 Mar 2. Biosens Bioelectron. 2021. PMID: 33706158 Free PMC article. Review.
-
Applications of gold nanoparticles in the detection and identification of infectious diseases and biothreats.Adv Mater. 2013 Jul 5;25(25):3490-6. doi: 10.1002/adma.201301333. Adv Mater. 2013. PMID: 23977699 Free PMC article.
-
Recent Developments in Nanotechnology-Based Biosensors for the Diagnosis of Coronavirus.Plasmonics. 2023;18(3):955-969. doi: 10.1007/s11468-023-01822-z. Epub 2023 Mar 27. Plasmonics. 2023. PMID: 37229148 Free PMC article. Review.
-
AuNPs for identification of molecular signatures of resistance.Front Microbiol. 2014 Aug 28;5:455. doi: 10.3389/fmicb.2014.00455. eCollection 2014. Front Microbiol. 2014. PMID: 25221547 Free PMC article. Review.
-
Mangiferin Against Respiratory Diseases: Pharmacological Targets and Prospects.Pharmacol Res Perspect. 2025 Aug;13(4):e70163. doi: 10.1002/prp2.70163. Pharmacol Res Perspect. 2025. PMID: 40783911 Free PMC article. Review.
References
FURTHER READING
-
- Driskell J, Kwarta K, Lipert R, Porter M, Neill J, Ridpath J. Low‐Level Detection of Viral Pathogens by a Surface‐Enhanced Raman Scattering Based Immunoassay. Anal Chem 2005, 77:6147–6154. - PubMed
-
- Fatemi K, Ghourchian H, Ziaee A‐A, Samiei S, Hanaee H. Paramagnetic nanoparticle‐based detection of hepatitis B virus using cathodic stripping voltammetry. Biotechnol Appl Biochem 2009, 052:221–225. - PubMed
-
- Fuentes M, Mateo C, Rodriguez A, Casqueiro M, Tercero J, Riese H, Fernandez‐Lafuente R, Guisan JM. Detecting minimal traces of DNA using DNA covalently attached to superparamagnetic nanoparticles and direct PCR‐ELISA. Biosens Bioelectron 2006, 21:1574‐1580. - PubMed
-
- Gao J, Gu H, Xu B. Multifunctional Magnetic Nanoparticles: Design, Synthesis, and Biomedical Applications. Acc Chem Res 2009, 42:1097–1107. - PubMed
References
-
- World Health Organization. Influenze WHO Fact Sheet no. 211, 2003.
-
- Collins PL, Crowe JE Jr. Respiratory syncytial virus and metapneumovirus In: David PH, Knipe M, eds. Fields Virology, vol. 2. Philadelphia: Lippincott Williams & Wilkins; 2007, 1601–1646.
-
- Prieto C, Castro JM. Porcine reproductive and respiratory syndrome virus infection in the boar: a review. Theriogenology 2005, 63: 1–16. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

