Structure of the 1,N(2)-etheno-2'-deoxyguanosine lesion in the 3'-G(epsilon dG)T-5' sequence opposite a one-base deletion
- PMID: 20201499
- PMCID: PMC2844103
- DOI: 10.1021/bi901516d
Structure of the 1,N(2)-etheno-2'-deoxyguanosine lesion in the 3'-G(epsilon dG)T-5' sequence opposite a one-base deletion
Abstract
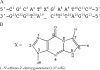
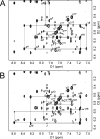
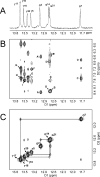
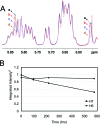
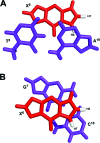
The structure of the 1,N(2)-ethenodeoxyguanosine lesion (1,N(2)-epsilondG) has been characterized in 5'-d(CGCATXGAATCC)-3'.5'-d(GGATTCATGCG)-3' (X = 1,N(2)-epsilondG), in which there is no dC opposite the lesion. This duplex (named the 1-BD duplex) models the product of translesion bypass of 1,N(2)-epsilondG by Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4) [Zang, H., Goodenough, A. K., Choi, J. Y., Irimia, A., Loukachevitch, L. V., Kozekov, I. D., Angel, K. C., Rizzo, C. J., Egli, M., and Guengerich, F. P. (2005) J. Biol. Chem. 280, 29750-29764], leading to a one-base deletion. The T(m) of this duplex is 6 degrees C higher than that of the duplex in which dC is present opposite the 1,N(2)-epsilondG lesion and 8 degrees C higher than that of the unmodified 1-BD duplex. Analysis of NOEs between the 1,N(2)-epsilondG imidazole and deoxyribose H1' protons and between the 1,N(2)-epsilondG etheno H6 and H7 protons and DNA protons establishes that 1,N(2)-epsilondG adopts the anti conformation about the glycosyl bond and that the etheno moiety is accommodated within the helix. The resonances of the 1,N(2)-epsilondG H6 and H7 etheno protons shift upfield relative to the monomer 1,N(2)-epsilondG, attributed to ring current shielding, consistent with their intrahelical location. NMR data reveal that Watson-Crick base pairing is maintained at both the 5' and 3' neighbor base pairs. The structure of the 1-BD duplex has been refined using molecular dynamics calculations restrained by NMR-derived distance and dihedral angle restraints. The increased stability of the 1,N(2)-epsilondG lesion in the absence of the complementary dC correlates with the one-base deletion extension product observed during the bypass of the 1,N(2)-epsilondG lesion by the Dpo4 polymerase, suggesting that stabilization of this bulged intermediate may be significant with regard to the biological processing of the lesion.
Figures






References
-
- Barbin A. (2000) Etheno-adduct-forming chemicals: From mutagenicity testing to tumor mutation spectra. Mutat. Res. 462, 55–69. - PubMed
-
- Barbin A.; Bresil H.; Croisy A.; Jacquignon P.; Malaveille C.; Montesano R.; Bartsch H. (1975) Liver-microsome-mediated formation of alkylating agents from vinyl bromide and vinyl chloride. Biochem. Biophys. Res. Commun. 67, 596–603. - PubMed
-
- Green T.; Hathway D. E. (1978) Interactions of vinyl chloride with rat-liver DNA in vivo. Chem.-Biol. Interact. 22, 211–224. - PubMed
-
- Eberle G.; Barbin A.; Laib R. J.; Ciroussel F.; Thomale J.; Bartsch H.; Rajewsky M. F. (1989) 1,N6-Etheno-2′-deoxyadenosine and 3,N4-etheno-2′-deoxycytidine detected by monoclonal antibodies in lung and liver DNA of rats exposed to vinyl chloride. Carcinogenesis 10, 209–212. - PubMed
-
- Fedtke N.; Boucheron J. A.; Walker V. E.; Swenberg J. A. (1990) Vinyl chloride-induced DNA adducts. II: Formation and persistence of 7-(2′-oxoethyl)guanine and N2,3-ethenoguanine in rat tissue DNA. Carcinogenesis 11, 1287–1292. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

