Genome dynamics of Bartonella grahamii in micro-populations of woodland rodents
- PMID: 20202191
- PMCID: PMC2847970
- DOI: 10.1186/1471-2164-11-152
Genome dynamics of Bartonella grahamii in micro-populations of woodland rodents
Abstract
Background: Rodents represent a high-risk reservoir for the emergence of new human pathogens. The recent completion of the 2.3 Mb genome of Bartonella grahamii, one of the most prevalent blood-borne bacteria in wild rodents, revealed a higher abundance of genes for host-cell interaction systems than in the genomes of closely related human pathogens. The sequence variability within the global B. grahamii population was recently investigated by multi locus sequence typing, but no study on the variability of putative host-cell interaction systems has been performed.
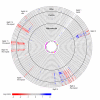
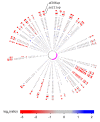
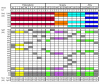
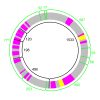
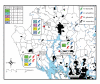
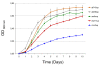
Results: To study the population dynamics of B. grahamii, we analyzed the genomic diversity on a whole-genome scale of 27 B. grahamii strains isolated from four different species of wild rodents in three geographic locations separated by less than 30 km. Even using highly variable spacer regions, only 3 sequence types were identified. This low sequence diversity contrasted with a high variability in genome content. Microarray comparative genome hybridizations identified genes for outer surface proteins, including a repeated region containing the fha gene for filamentous hemaggluttinin and a plasmid that encodes a type IV secretion system, as the most variable. The estimated generation times in liquid culture medium for a subset of strains ranged from 5 to 22 hours, but did not correlate with sequence type or presence/absence patterns of the fha gene or the plasmid.
Conclusion: Our study has revealed a geographic microstructure of B. grahamii in wild rodents. Despite near-identity in nucleotide sequence, major differences were observed in gene presence/absence patterns that did not segregate with host species. This suggests that genetically similar strains can infect a range of different hosts.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

