Cell cycle-dependent regulation of the bi-directional overlapping promoter of human BRCA2/ZAR2 genes in breast cancer cells
- PMID: 20202217
- PMCID: PMC2842238
- DOI: 10.1186/1476-4598-9-50
Cell cycle-dependent regulation of the bi-directional overlapping promoter of human BRCA2/ZAR2 genes in breast cancer cells
Abstract
Background: BRCA2 gene expression is tightly regulated during the cell cycle in human breast cells. The expression of BRCA2 gene is silenced at the G0/G1 phase of cell growth and is de-silenced at the S/G2 phase. While studying the activity of BRCA2 gene promoter in breast cancer cells, we discovered that this promoter has bi-directional activity and the product of the reverse activity (a ZAR1-like protein, we named ZAR2) silences the forward promoter at the G0/G1 phase of the cell. Standard techniques like cell synchronization by serum starvation, flow cytometry, N-terminal or C-terminal FLAG epitope-tagged protein expression, immunofluorescence confocal microscopy, dual luciferase assay for promoter evaluation, and chromatin immunoprecipitation assay were employed during this study.
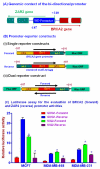
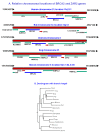
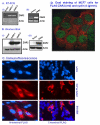
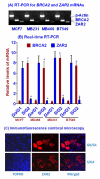
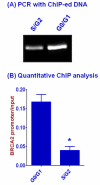
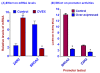
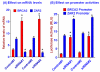
Results: Human BRCA2 gene promoter is active in both the forward and the reverse orientations. This promoter is 8-20 fold more active in the reverse orientation than in the forward orientation when the cells are in the non-dividing stage (G0/G1). When the cells are in the dividing state (S/G2), the forward activity of the promoter is 5-8 folds higher than the reverse activity. The reverse activity transcribes the ZAR2 mRNA with 966 nt coding sequence which codes for a 321 amino acid protein. ZAR2 has two C4 type zinc fingers at the carboxyl terminus. In the G0/G1 growth phase ZAR2 is predominantly located inside the nucleus of the breast cells, binds to the BRCA2 promoter and inhibits the expression of BRCA2. In the dividing cells, ZAR2 is trapped in the cytoplasm.
Conclusions: BRCA2 gene promoter has bi-directional activity, expressing BRCA2 and a novel C4-type zinc finger containing transcription factor ZAR2. Subcellular location of ZAR2 and its expression from the reverse promoter of the BRCA2 gene are stringently regulated in a cell cycle dependent manner. ZAR2 binds to BRCA2/ZAR2 bi-directional promoter in vivo and is responsible, at least in part, for the silencing of BRCA2 gene expression in the G0/G1 phase in human breast cells.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

