Collective mass spectrometry approaches reveal broad and combinatorial modification of high mobility group protein A1a
- PMID: 20202861
- PMCID: PMC3321734
- DOI: 10.1016/j.jasms.2010.01.020
Collective mass spectrometry approaches reveal broad and combinatorial modification of high mobility group protein A1a
Abstract
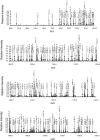
Transcriptional states are formed and maintained by the interaction and post-translational modification (PTM) of several chromatin proteins, such as histones and high mobility group (HMG) proteins. Among these, HMGA1a, a small heterochromatin-associated nuclear protein has been shown to be post-translationally modified, and some of these PTMs have been linked to apoptosis and cancer. In cancerous cells, HMGA1a PTMs differ between metastatic and nonmetastatic cells, suggesting the existence of an HMGA1a PTM code analogous to the "histone code." In this study, we expand on current knowledge by comprehensively characterizing PTMs on HMGA1a purified from human cells using both nanoflow liquid chromatography collision activated dissociation mediated Bottom Up and electron-transfer dissociation facilitated middle and Top Down mass spectrometry (MS). We find HMGA1a to be pervasively modified with many types of modifications such as methylation, acetylation, and phosphorylation, including finding novel sites. While Bottom Up MS identified lower level modification sites, Top and Middle Down MS were utilized to identify the most commonly occurring combinatorially modified forms. Remarkably, although we identify several individual modification sites through our Bottom Up and Middle Down MS analyses, we find relatively few combinatorially modified forms dominate the population through Top Down proteomics. The main combinatorial PTMs we find through the Top Down approach are N-terminal acetylation, Arg25 methylation along with phosphorylation of the three most C-terminal serine residues in primarily a diphosphorylated form. This report presents one of the most detailed analyses of HMGA1a to date and illustrates the strength of using a combined MS effort.
Copyright 2010. Published by Elsevier Inc.
Figures




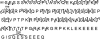
Similar articles
-
In vivo posttranslational modifications of the high mobility group A1a proteins in breast cancer cells of differing metastatic potential.Biochemistry. 2004 Sep 14;43(36):11500-15. doi: 10.1021/bi049833i. Biochemistry. 2004. PMID: 15350136
-
Dynamic and differential in vivo modifications of the isoform HMGA1a and HMGA1b chromatin proteins.J Biol Chem. 2005 Mar 11;280(10):8961-73. doi: 10.1074/jbc.M407348200. Epub 2004 Dec 11. J Biol Chem. 2005. PMID: 15591590
-
Tandem mass spectrometry for the examination of the posttranslational modifications of high-mobility group A1 proteins: symmetric and asymmetric dimethylation of Arg25 in HMGA1a protein.Biochemistry. 2005 Apr 26;44(16):6293-301. doi: 10.1021/bi0475525. Biochemistry. 2005. PMID: 15835918
-
Combinations of histone post-translational modifications.Biochem J. 2021 Feb 12;478(3):511-532. doi: 10.1042/BCJ20200170. Biochem J. 2021. PMID: 33567070 Review.
-
The top-down, middle-down, and bottom-up mass spectrometry approaches for characterization of histone variants and their post-translational modifications.Proteomics. 2014 Mar;14(4-5):489-97. doi: 10.1002/pmic.201300256. Epub 2013 Dec 16. Proteomics. 2014. PMID: 24339419 Review.
Cited by
-
Middle-Down and Top-Down Mass Spectrometric Analysis of Co-occurring Histone Modifications.Curr Protoc Protein Sci. 2014 Aug 1;77:23.7.1-23.7.28. doi: 10.1002/0471140864.ps2307s77. Curr Protoc Protein Sci. 2014. PMID: 25081742 Free PMC article. Review.
-
Characterization of green fluorescent proteins by 193 nm ultraviolet photodissociation mass spectrometry.Proteomics. 2014 May;14(10):1165-73. doi: 10.1002/pmic.201300364. Epub 2014 Apr 10. Proteomics. 2014. PMID: 24596159 Free PMC article.
-
Proteomics in epigenetics: new perspectives for cancer research.Brief Funct Genomics. 2013 May;12(3):205-18. doi: 10.1093/bfgp/elt002. Epub 2013 Feb 11. Brief Funct Genomics. 2013. PMID: 23401080 Free PMC article. Review.
-
Apelin-mediated deamidation of HMGA1 promotes tumorigenesis by enhancing SREBP1 activity and lipid synthesis.Cancer Sci. 2022 Nov;113(11):3722-3734. doi: 10.1111/cas.15515. Epub 2022 Sep 10. Cancer Sci. 2022. PMID: 36087034 Free PMC article.
-
Analysis of intact protein isoforms by mass spectrometry.J Biol Chem. 2011 Jul 22;286(29):25451-8. doi: 10.1074/jbc.R111.239442. Epub 2011 Jun 1. J Biol Chem. 2011. PMID: 21632550 Free PMC article. Review.
References
-
- Bernstein BE, Meissner A, Lander ES. The Mammalian Epigenome. Cell. 2007;128:669–681. - PubMed
-
- Cheung P, Allis CD, Sassone-Corsi P. Signaling to Chromatin through Histone Modifications. Cell. 2000;103:263–271. - PubMed
-
- Kouzarides T. Chromatin Modifications and Their Function. Cell. 2007;128:693–705. - PubMed
-
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of Histone H3 Lysine 9 Creates a Binding Site for Hp1 Proteins. Nature. 2001;410:116–120. - PubMed
-
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective Recognition of Methylated Lysine 9 on Histone H3 by the Hp1 Chromo Domain. Nature. 2001;410:120–124. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

