Mapping of domains on HIV envelope protein mediating association with calnexin and protein-disulfide isomerase
- PMID: 20202930
- PMCID: PMC2859542
- DOI: 10.1074/jbc.M109.066670
Mapping of domains on HIV envelope protein mediating association with calnexin and protein-disulfide isomerase
Abstract
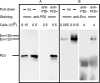
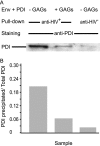
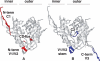
The cell catalysts calnexin (CNX) and protein-disulfide isomerase (PDI) cooperate in establishing the disulfide bonding of the HIV envelope (Env) glycoprotein. Following HIV binding to lymphocytes, cell-surface PDI also reduces Env to induce the fusogenic conformation. We sought to define the contact points between Env and these catalysts to illustrate their potential as therapeutic targets. In lysates of Env-expressing cells, 15% of the gp160 precursor, but not gp120, coprecipitated with CNX, whereas only 0.25% of gp160 and gp120 coprecipitated with PDI. Under in vitro conditions, which mimic the Env/PDI interaction during virus/cell contact, PDI readily associated with Env. The domains of Env interacting in cellulo with CNX or in vitro with PDI were then determined using anti-Env antibodies whose binding site was occluded by CNX or PDI. Antibodies against domains V1/V2, C2, and the C terminus of V3 did not bind CNX-associated Env, whereas those against C1, V1/V2, and the CD4-binding domain did not react with PDI-associated Env. In addition, a mixture of the latter antibodies interfered with PDI-mediated Env reduction. Thus, Env interacts with intracellular CNX and extracellular PDI via discrete, largely nonoverlapping, regions. The sites of interaction explain the mode of action of compounds that target these two catalysts and may enable the design of further new competitive agents.
Figures


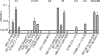


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

