Genomic structure of an economically important cyanobacterium, Arthrospira (Spirulina) platensis NIES-39
- PMID: 20203057
- PMCID: PMC2853384
- DOI: 10.1093/dnares/dsq004
Genomic structure of an economically important cyanobacterium, Arthrospira (Spirulina) platensis NIES-39
Abstract
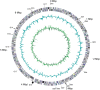
A filamentous non-N(2)-fixing cyanobacterium, Arthrospira (Spirulina) platensis, is an important organism for industrial applications and as a food supply. Almost the complete genome of A. platensis NIES-39 was determined in this study. The genome structure of A. platensis is estimated to be a single, circular chromosome of 6.8 Mb, based on optical mapping. Annotation of this 6.7 Mb sequence yielded 6630 protein-coding genes as well as two sets of rRNA genes and 40 tRNA genes. Of the protein-coding genes, 78% are similar to those of other organisms; the remaining 22% are currently unknown. A total 612 kb of the genome comprise group II introns, insertion sequences and some repetitive elements. Group I introns are located in a protein-coding region. Abundant restriction-modification systems were determined. Unique features in the gene composition were noted, particularly in a large number of genes for adenylate cyclase and haemolysin-like Ca(2+)-binding proteins and in chemotaxis proteins. Filament-specific genes were highlighted by comparative genomic analysis.
Figures


Similar articles
-
Whole genomic DNA sequencing and comparative genomic analysis of Arthrospira platensis: high genome plasticity and genetic diversity.DNA Res. 2016 Aug;23(4):325-38. doi: 10.1093/dnares/dsw023. Epub 2016 Jun 21. DNA Res. 2016. PMID: 27330141 Free PMC article.
-
The Draft Genome of a Hydrogen-producing Cyanobacterium, Arthrospira platensis NIES-46.J Genomics. 2019 Sep 18;7:56-59. doi: 10.7150/jgen.38149. eCollection 2019. J Genomics. 2019. PMID: 31588248 Free PMC article.
-
The AplI restriction-modification system in an edible cyanobacterium, Arthrospira (Spirulina) platensis NIES-39, recognizes the nucleotide sequence 5'-CTGCAG-3'.Biosci Biotechnol Biochem. 2013;77(4):782-8. doi: 10.1271/bbb.120919. Epub 2013 Apr 7. Biosci Biotechnol Biochem. 2013. PMID: 23563565
-
Lipophilic and Hydrophilic Compounds from Arthrospira platensis and Its Effects on Tissue and Blood Cells-An Overview.Life (Basel). 2022 Sep 26;12(10):1497. doi: 10.3390/life12101497. Life (Basel). 2022. PMID: 36294932 Free PMC article. Review.
-
Application of Arthrospira platensis for Medicinal Purposes and the Food Industry: A Review of the Literature.Life (Basel). 2023 Mar 21;13(3):845. doi: 10.3390/life13030845. Life (Basel). 2023. PMID: 36984000 Free PMC article. Review.
Cited by
-
Molecular investigation of the radiation resistance of edible cyanobacterium Arthrospira sp. PCC 8005.Microbiologyopen. 2015 Apr;4(2):187-207. doi: 10.1002/mbo3.229. Epub 2015 Feb 12. Microbiologyopen. 2015. PMID: 25678338 Free PMC article.
-
A pilot-scale floating closed culture system for the multicellular cyanobacterium Arthrospira platensis NIES-39.J Appl Phycol. 2015;27(6):2191-2202. doi: 10.1007/s10811-014-0484-2. Epub 2014 Dec 8. J Appl Phycol. 2015. PMID: 26523091 Free PMC article.
-
Emerging Species and Genome Editing Tools: Future Prospects in Cyanobacterial Synthetic Biology.Microorganisms. 2019 Sep 29;7(10):409. doi: 10.3390/microorganisms7100409. Microorganisms. 2019. PMID: 31569579 Free PMC article. Review.
-
Bacterial group I introns: mobile RNA catalysts.Mob DNA. 2014 Mar 10;5(1):8. doi: 10.1186/1759-8753-5-8. Mob DNA. 2014. PMID: 24612670 Free PMC article.
-
Whole genomic DNA sequencing and comparative genomic analysis of Arthrospira platensis: high genome plasticity and genetic diversity.DNA Res. 2016 Aug;23(4):325-38. doi: 10.1093/dnares/dsw023. Epub 2016 Jun 21. DNA Res. 2016. PMID: 27330141 Free PMC article.
References
-
- Desikachary T.V. In: Botanical Monographs. Carr N.G., Whitton B.A., editors. Oxford, London, Edinburgh, Melbourne: Blackwell Scientific Publications; 1973. pp. 473–81.
-
- Doolittle W.F. In: The Biology of Cyanobacteria, Botanical Monographs. Carr N.G., Whitton B.A., editors. Berkeley and Los Angels: University of California Press; 1982. pp. 307–31.
-
- Van Tuerenhout D. The Aztecs: New Perspectives. ABC-CLIO; 2005.
-
- Belay A. In: Spirulina in Human Nutrition and Health. Gershwin M.E., Belay A., editors. CRC press: Florida; 2007. pp. 1–26.
-
- Castenholz R.W., Rippka R., Herdman M., Wilmotte A. In: Bergey's Manual of Systematic Bacteriology. 2nd edition. Boone D.R., Castenholz R.W., Garrity G.M., editors. Springer: Berlin; 2007. pp. 542–3.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

