The crystal structure of unmodified tRNAPhe from Escherichia coli
- PMID: 20203084
- PMCID: PMC2896525
- DOI: 10.1093/nar/gkq133
The crystal structure of unmodified tRNAPhe from Escherichia coli
Abstract
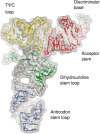
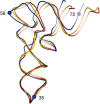
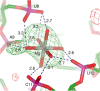
Post-transcriptional nucleoside modifications fine-tune the biophysical and biochemical properties of transfer RNA (tRNA) so that it is optimized for participation in cellular processes. Here we report the crystal structure of unmodified tRNA(Phe) from Escherichia coli at a resolution of 3 A. We show that in the absence of modifications the overall fold of the tRNA is essentially the same as that of mature tRNA. However, there are a number of significant structural differences, such as rearrangements in a triplet base pair and a widened angle between the acceptor and anticodon stems. Contrary to previous observations, the anticodon adopts the same conformation as seen in mature tRNA.
Figures

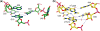
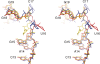

References
-
- Schurer H, Schiffer S, Marchfelder A, Morl M. This is the end: processing, editing and repair at the tRNA 3′-terminus. Biol. Chem. 2001;382:1147–1156. - PubMed
-
- Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, Clark BFC, Klug A. Structure of yeast phenylalanine transfer RNA at 3 Å resolution. Nature. 1974;250:546–551. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions

