GEN1/Yen1 and the SLX4 complex: Solutions to the problem of Holliday junction resolution
- PMID: 20203129
- PMCID: PMC2841330
- DOI: 10.1101/gad.1903510
GEN1/Yen1 and the SLX4 complex: Solutions to the problem of Holliday junction resolution
Abstract
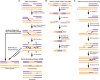
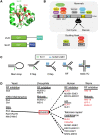
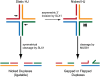
Chromosomal double-strand breaks (DSBs) are considered to be among the most deleterious DNA lesions found in eukaryotic cells due to their propensity to promote genome instability. DSBs occur as a result of exogenous or endogenous DNA damage, and also occur during meiotic recombination. DSBs are often repaired through a process called homologous recombination (HR), which employs the sister chromatid in mitotic cells or the homologous chromosome in meiotic cells, as a template for repair. HR frequently involves the formation and resolution of four-way DNA structures referred to as the Holliday junction (HJ). Despite extensive study, the machinery and mechanisms used to process these structures in eukaryotes have remained poorly understood. Recent work has identified XPG and UvrC/GIY domain-containing structure-specific endonucleases that can symmetrically cleave HJs in vitro in a manner that allows for religation without additional processing, properties that are reminiscent of the classical RuvC HJ resolvase in bacteria. Genetic studies reveal potential roles for these HJ resolvases in repair after DNA damage and during meiosis. The stage is now set for a more comprehensive understanding of the specific roles these enzymes play in the response of cells to DSBs, collapsed replication forks, telomere dysfunction, and meiotic recombination.
Figures




References
-
- Allers T, Lichten M. Intermediates of yeast meiotic recombination contain heteroduplex DNA. Mol Cell. 2001;8:225–231. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
