Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom)
- PMID: 20203130
- PMCID: PMC2841335
- DOI: 10.1101/gad.1898410
Linking H3K79 trimethylation to Wnt signaling through a novel Dot1-containing complex (DotCom)
Abstract
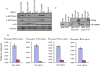
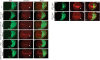
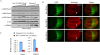
Epigenetic modifications of chromatin play an important role in the regulation of gene expression. KMT4/Dot1 is a conserved histone methyltransferase capable of methylating chromatin on Lys79 of histone H3 (H3K79). Here we report the identification of a multisubunit Dot1 complex (DotCom), which includes several of the mixed lineage leukemia (MLL) partners in leukemia such as ENL, AF9/MLLT3, AF17/MLLT6, and AF10/MLLT10, as well as the known Wnt pathway modifiers TRRAP, Skp1, and beta-catenin. We demonstrated that the human DotCom is indeed capable of trimethylating H3K79 and, given the association of beta-catenin, Skp1, and TRRAP, we investigated, and found, a role for Dot1 in Wnt/Wingless signaling in an in vivo model system. Knockdown of Dot1 in Drosophila results in decreased expression of a subset of Wingless target genes. Furthermore, the loss of expression for the Drosophila homologs of the Dot1-associated proteins involved in the regulation of H3K79 shows a similar reduction in expression of these Wingless targets. From yeast to human, specific trimethylation of H3K79 by Dot1 requires the monoubiquitination of histone H2B by the Rad6/Bre1 complex. Here, we demonstrate that depletion of Bre1, the E3 ligase required for H2B monoubiquitination, leads specifically to reduced bulk H3K79 trimethylation levels and a reduction in expression of many Wingless targets. Overall, our study describes for the first time the components of DotCom and links the specific regulation of H3K79 trimethylation by Dot1 and its associated factors to the Wnt/Wingless signaling pathway.
Figures



References
-
- Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat Struct Mol Biol. 2007;14:1008–1016. - PubMed
-
- Bitoun E, Oliver PL, Davies KE. The mixed-lineage leukemia fusion partner AF4 stimulates RNA polymerase II transcriptional elongation and mediates coordinated chromatin remodeling. Hum Mol Genet. 2007;16:92–106. - PubMed
-
- Bray S, Musisi H, Bienz M. Bre1 is required for Notch signaling and histone modification. Dev Cell. 2005;8:279–286. - PubMed
-
- Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD. Gene silencing: Trans-histone regulatory pathway in chromatin. Nature. 2002;418:498. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
