Phylogeography takes a relaxed random walk in continuous space and time
- PMID: 20203288
- PMCID: PMC2915639
- DOI: 10.1093/molbev/msq067
Phylogeography takes a relaxed random walk in continuous space and time
Abstract
Research aimed at understanding the geographic context of evolutionary histories is burgeoning across biological disciplines. Recent endeavors attempt to interpret contemporaneous genetic variation in the light of increasingly detailed geographical and environmental observations. Such interest has promoted the development of phylogeographic inference techniques that explicitly aim to integrate such heterogeneous data. One promising development involves reconstructing phylogeographic history on a continuous landscape. Here, we present a Bayesian statistical approach to infer continuous phylogeographic diffusion using random walk models while simultaneously reconstructing the evolutionary history in time from molecular sequence data. Moreover, by accommodating branch-specific variation in dispersal rates, we relax the most restrictive assumption of the standard Brownian diffusion process and demonstrate increased statistical efficiency in spatial reconstructions of overdispersed random walks by analyzing both simulated and real viral genetic data. We further illustrate how drawing inference about summary statistics from a fully specified stochastic process over both sequence evolution and spatial movement reveals important characteristics of a rabies epidemic. Together with recent advances in discrete phylogeographic inference, the continuous model developments furnish a flexible statistical framework for biogeographical reconstructions that is easily expanded upon to accommodate various landscape genetic features.
Figures

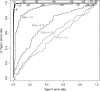
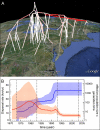
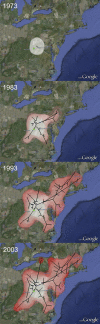
References
-
- Andrews D, Mallows C. Scale mixtures of normal distributions. J R Stat Soc Series B. 1974;36:99–102.
-
- Berger J, Jefferys W. Ockham's razor and Bayesian analysis. Am Sci. 1992;80:64–72.
-
- Brown R. A brief account of microscopical observations made in the months of June, July and August, 1827, on the particles contained in the pollen of plants; and on the general existence of active molecules in organic and inorganic bodies. Philos Mag. 1828;4:161–173.
-
- Butler M, King A. Phylogenetic comparative analysis: a modeling approach for adaptive evolution. Am Nat. 2004;164:683–695. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

