The mycobacterial Mpa-proteasome unfolds and degrades pupylated substrates by engaging Pup's N-terminus
- PMID: 20203624
- PMCID: PMC2857465
- DOI: 10.1038/emboj.2010.23
The mycobacterial Mpa-proteasome unfolds and degrades pupylated substrates by engaging Pup's N-terminus
Abstract
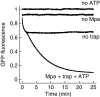
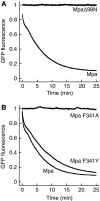
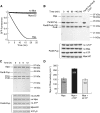
Mycobacterium tuberculosis, along with other actinobacteria, harbours proteasomes in addition to members of the general bacterial repertoire of degradation complexes. In analogy to ubiquitination in eukaryotes, substrates are tagged for proteasomal degradation with prokaryotic ubiquitin-like protein (Pup) that is recognized by the N-terminal coiled-coil domain of the ATPase Mpa (also called ARC). Here, we reconstitute the entire mycobacterial proteasome degradation system for pupylated substrates and establish its mechanistic features with respect to substrate recruitment, unfolding and degradation. We show that the Mpa-proteasome complex unfolds and degrades Pup-tagged proteins and that this activity requires physical interaction of the ATPase with the proteasome. Furthermore, we establish the N-terminal region of Pup as the structural element required for engagement of pupylated substrates into the Mpa pore. In this process, Mpa pulls on Pup to initiate unfolding of substrate proteins and to drag them toward the proteasome chamber. Unlike the eukaryotic ubiquitin, Pup is not recycled but degraded with the substrate. This assigns a dual function to Pup as both the Mpa recognition element as well as the threading determinant.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

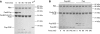

Comment in
-
Pup grows up: in vitro characterization of the degradation of pupylated proteins.EMBO J. 2010 Apr 7;29(7):1163-4. doi: 10.1038/emboj.2010.40. EMBO J. 2010. PMID: 20372178 Free PMC article.
References
-
- Chaudhuri BN, Sawaya MR, Kim CY, Waldo GS, Park MS, Terwilliger TC, Yeates TO (2003) The crystal structure of the first enzyme in the pantothenate biosynthetic pathway, ketopantoate hydroxymethyltransferase, from M tuberculosis. Structure 11: 753–764 - PubMed
-
- Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW, Gygi SP, Finley D (2006) Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell 127: 1401–1413 - PubMed
-
- Darwin KH, Ehrt S, Gutierrez-Ramos JC, Weich N, Nathan CF (2003) The proteasome of Mycobacterium tuberculosis is required for resistance to nitric oxide. Science 302: 1963–1966 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

