Evolution and diversity of the human hepatitis d virus genome
- PMID: 20204073
- PMCID: PMC2829689
- DOI: 10.1155/2010/323654
Evolution and diversity of the human hepatitis d virus genome
Abstract
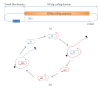
Human hepatitis delta virus (HDV) is the smallest RNA virus in genome. HDV genome is divided into a viroid-like sequence and a protein-coding sequence which could have originated from different resources and the HDV genome was eventually constituted through RNA recombination. The genome subsequently diversified through accumulation of mutations selected by interactions between the mutated RNA and proteins with host factors to successfully form the infectious virions. Therefore, we propose that the conservation of HDV nucleotide sequence is highly related with its functionality. Genome analysis of known HDV isolates shows that the C-terminal coding sequences of large delta antigen (LDAg) are the highest diversity than other regions of protein-coding sequences but they still retain biological functionality to interact with the heavy chain of clathrin can be selected and maintained. Since viruses interact with many host factors, including escaping the host immune response, how to design a program to predict RNA genome evolution is a great challenging work.
Figures
References
-
- Chen PJ, Chen DS. Hepatitis B virus infection. The New England Journal of Medicine. 1998;338:1312–1313. - PubMed
-
- Tiollais P, Pourcel C, Dejean A. The hepatitis B virus. Nature. 1985;317:489–495. - PubMed
-
- Gust ID, Burrell CJ, Coulepis AG, Robinson WS, Zuckerman AJ. Taxonomic classification of human hepatitis B virus. Intervirology. 1986;25(1):14–29. - PubMed
-
- Lai MMC. The molecular biology of hepatitis delta virus. Annual Review of Biochemistry. 1995;64:259–286. - PubMed
-
- Makino S, Chang M-F, Shieh C-K, et al. Molecular cloning and sequencing of a human hepatitis delta (δ) virus RNA. Nature. 1987;329(6137):343–346. - PubMed
LinkOut - more resources
Full Text Sources

