The Sinorhizobium meliloti RNA chaperone Hfq influences central carbon metabolism and the symbiotic interaction with alfalfa
- PMID: 20205931
- PMCID: PMC2848018
- DOI: 10.1186/1471-2180-10-71
The Sinorhizobium meliloti RNA chaperone Hfq influences central carbon metabolism and the symbiotic interaction with alfalfa
Abstract
Background: The bacterial Hfq protein is able to interact with diverse RNA molecules, including regulatory small non-coding RNAs (sRNAs), and thus it is recognized as a global post-transcriptional regulator of gene expression. Loss of Hfq has an extensive impact in bacterial physiology which in several animal pathogens influences virulence. Sinorhizobium meliloti is a model soil bacterium known for its ability to establish a beneficial nitrogen-fixing intracellular symbiosis with alfalfa. Despite the predicted general involvement of Hfq in the establishment of successful bacteria-eukaryote interactions, its function in S. meliloti has remained unexplored.
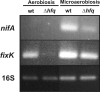
Results: Two independent S. meliloti mutants, 2011-3.4 and 1021Deltahfq, were obtained by disruption and deletion of the hfq gene in the wild-type strains 2011 and 1021, respectively, both exhibiting similar growth defects as free-living bacteria. Transcriptomic profiling of 1021Deltahfq revealed a general down-regulation of genes of sugar transporters and some enzymes of the central carbon metabolism, whereas transcripts specifying the uptake and metabolism of nitrogen sources (mainly amino acids) were more abundant than in the wild-type strain. Proteomic analysis of the 2011-3.4 mutant independently confirmed these observations. Symbiotic tests showed that lack of Hfq led to a delayed nodulation, severely compromised bacterial competitiveness on alfalfa roots and impaired normal plant growth. Furthermore, a large proportion of nodules (55%-64%) elicited by the 1021Deltahfq mutant were non-fixing, with scarce content in bacteroids and signs of premature senescence of endosymbiotic bacteria. RT-PCR experiments on RNA from bacteria grown under aerobic and microoxic conditions revealed that Hfq contributes to regulation of nifA and fixK1/K2, the genes controlling nitrogen fixation, although the Hfq-mediated regulation of fixK is only aerobiosis dependent. Finally, we found that some of the recently identified S. meliloti sRNAs co-inmunoprecipitate with a FLAG-epitope tagged Hfq protein.
Conclusions: Our results support that the S. meliloti RNA chaperone Hfq contributes to the control of central metabolic pathways in free-living bacteria and influences rhizospheric competence, survival of the microsymbiont within the nodule cells and nitrogen fixation during the symbiotic interaction with its legume host alfalfa. The identified S. meliloti Hfq-binding sRNAs are predicted to participate in the Hfq regulatory network.
Figures





Similar articles
-
The Sinorhizobium meliloti RNA chaperone Hfq mediates symbiosis of S. meliloti and alfalfa.J Bacteriol. 2010 Mar;192(6):1710-8. doi: 10.1128/JB.01427-09. Epub 2010 Jan 15. J Bacteriol. 2010. PMID: 20081033 Free PMC article.
-
Genome-wide profiling of Hfq-binding RNAs uncovers extensive post-transcriptional rewiring of major stress response and symbiotic regulons in Sinorhizobium meliloti.RNA Biol. 2014;11(5):563-79. doi: 10.4161/rna.28239. Epub 2014 Feb 26. RNA Biol. 2014. PMID: 24786641 Free PMC article.
-
Insights into the noncoding RNome of nitrogen-fixing endosymbiotic α-proteobacteria.Mol Plant Microbe Interact. 2013 Feb;26(2):160-7. doi: 10.1094/MPMI-07-12-0186-CR. Mol Plant Microbe Interact. 2013. PMID: 22991999 Review.
-
Contributions of Sinorhizobium meliloti Transcriptional Regulator DksA to Bacterial Growth and Efficient Symbiosis with Medicago sativa.J Bacteriol. 2016 Apr 14;198(9):1374-83. doi: 10.1128/JB.00013-16. Print 2016 May. J Bacteriol. 2016. PMID: 26883825 Free PMC article.
-
How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model.Nat Rev Microbiol. 2007 Aug;5(8):619-33. doi: 10.1038/nrmicro1705. Nat Rev Microbiol. 2007. PMID: 17632573 Free PMC article. Review.
Cited by
-
Analyzing the Complex Regulatory Landscape of Hfq - an Integrative, Multi-Omics Approach.Front Microbiol. 2017 Sep 20;8:1784. doi: 10.3389/fmicb.2017.01784. eCollection 2017. Front Microbiol. 2017. PMID: 29033902 Free PMC article.
-
Independent activity of the homologous small regulatory RNAs AbcR1 and AbcR2 in the legume symbiont Sinorhizobium meliloti.PLoS One. 2013 Jul 15;8(7):e68147. doi: 10.1371/journal.pone.0068147. Print 2013. PLoS One. 2013. PMID: 23869210 Free PMC article.
-
Quantitative proteomic analysis of the Hfq-regulon in Sinorhizobium meliloti 2011.PLoS One. 2012;7(10):e48494. doi: 10.1371/journal.pone.0048494. Epub 2012 Oct 30. PLoS One. 2012. PMID: 23119037 Free PMC article.
-
Analysis of the small RNA P16/RgsA in the plant pathogen Pseudomonas syringae pv. tomato strain DC3000.Microbiology (Reading). 2013 Feb;159(Pt 2):296-306. doi: 10.1099/mic.0.063826-0. Epub 2012 Dec 20. Microbiology (Reading). 2013. PMID: 23258266 Free PMC article.
-
Hfq influences multiple transport systems and virulence in the plant pathogen Agrobacterium tumefaciens.J Bacteriol. 2012 Oct;194(19):5209-17. doi: 10.1128/JB.00510-12. Epub 2012 Jul 20. J Bacteriol. 2012. PMID: 22821981 Free PMC article.
References
-
- Franze de Fernández MT, Hayward WS, August JT. Bacterial proteins required for replication of phage Q ribonucleic acid. Purification and properties of host factor I, a ribonucleic acid binding protein. J Biol Chem. 1972;247(3):824–831. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

