Complete genome sequences of avian paramyxovirus serotype 6 prototype strain Hong Kong and a recent novel strain from Italy: evidence for the existence of subgroups within the serotype
- PMID: 20206652
- PMCID: PMC2859990
- DOI: 10.1016/j.virusres.2010.02.015
Complete genome sequences of avian paramyxovirus serotype 6 prototype strain Hong Kong and a recent novel strain from Italy: evidence for the existence of subgroups within the serotype
Abstract
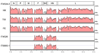
Complete genome sequences were determined for two strains of avian paramyxovirus serotype 6 (APMV-6): the prototype Hong Kong (HK) strain and a more recent isolate from Italy (IT4524-2). The genome length of strain HK is 16236 nucleotide (nt), which is the same as for the other two APMV-6 strains (FE and TW) that have been reported to date, whereas that of strain IT4524-2 is 16230 nt. The length difference in strain IT4524-2 is due to a 6-nt deletion in the downstream untranslated region of the F gene. All of these viruses follow the "rule of six". Each genome consists of seven genes in the order of 3'N-P-M-F-SH-HN-L5', which differs from other APMV serotypes in containing an additional gene encoding the small hydrophobic (SH) protein. Sequence comparisons revealed that strain IT4524-2 shares an unexpectedly low level of genome nt sequence identity (70%) and aggregate predicted amino acid (aa) sequence identity (79%) with other three strains, which in contrast are more closely related to each other with nt sequence 94-98% nt identity and 90-100% aggregate aa identity. Sequence analysis of the F-SH-HN genome region of two other recent Italian isolates showed that they fall in the HK/FE/TW group. The predicted signal peptide of IT4524-2 F protein lacks the N-terminal first 10 aa that are present in the other five strains. Also, the F protein cleavage site of strain IT4524-2, REPR downward arrow L, has two dibasic aa (arginine, R) compared to the monobasic F protein cleavage site of PEPR downward arrow L in the other strains. Reciprocal cross-hemagglutination inhibition (HI) assays using post-infection chicken sera indicated that strain IT4524-2 is antigenically related to the other APMV-6 strains, but with 4- to 8-fold lower HI tiers for the test sera between strain IT4524-2 and the other APMV-6 strains. Taken together, our results indicated that the APMV-6 strains represents a single serotype with two subgroups that differ substantially based on nt and aa sequences and can be distinguished by HI assay.
(c) 2010 Elsevier B.V. All rights reserved.
Figures






Similar articles
-
Complete genome sequence of avian paramyxovirus-3 strain Wisconsin: evidence for the existence of subgroups within the serotype.Virus Res. 2010 Apr;149(1):78-85. doi: 10.1016/j.virusres.2009.12.015. Epub 2010 Jan 15. Virus Res. 2010. PMID: 20079781 Free PMC article.
-
Complete genome sequences of avian paramyxovirus serotype 2 (APMV-2) strains Bangor, England and Kenya: evidence for the existence of subgroups within serotype 2.Virus Res. 2010 Sep;152(1-2):85-95. doi: 10.1016/j.virusres.2010.06.009. Epub 2010 Jun 22. Virus Res. 2010. PMID: 20600395 Free PMC article.
-
Complete genome sequence of avian paramyxovirus (APMV) serotype 5 completes the analysis of nine APMV serotypes and reveals the longest APMV genome.PLoS One. 2010 Feb 17;5(2):e9269. doi: 10.1371/journal.pone.0009269. PLoS One. 2010. PMID: 20174645 Free PMC article.
-
Molecular characterization and complete genome sequence of avian paramyxovirus type 4 prototype strain duck/Hong Kong/D3/75.Virol J. 2008 Oct 20;5:124. doi: 10.1186/1743-422X-5-124. Virol J. 2008. PMID: 18937854 Free PMC article.
-
Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited.J Virol. 1998 Feb;72(2):891-9. doi: 10.1128/JVI.72.2.891-899.1998. J Virol. 1998. PMID: 9444980 Free PMC article. Review. No abstract available.
Cited by
-
Characterization of novel avian paramyxovirus strain APMV/Shimane67 isolated from migratory wild geese in Japan.J Vet Med Sci. 2015 Sep;77(9):1079-85. doi: 10.1292/jvms.14-0529. Epub 2015 Apr 11. J Vet Med Sci. 2015. PMID: 25866408 Free PMC article.
-
Complete nucleotide sequence of avian paramyxovirus type 6 strain JL isolated from mallard ducks in China.J Virol. 2012 Dec;86(23):13112. doi: 10.1128/JVI.02317-12. J Virol. 2012. PMID: 23118446 Free PMC article.
-
Avian Paramyxovirus 4 Antitumor Activity Leads to Complete Remissions and Long-term Protective Memory in Preclinical Melanoma and Colon Carcinoma Models.Cancer Res Commun. 2022 Jul;2(7):602-615. doi: 10.1158/2767-9764.crc-22-0025. Epub 2022 Jul 7. Cancer Res Commun. 2022. PMID: 35937459 Free PMC article.
-
Identification and complete genome sequencing of paramyxoviruses in mallard ducks (Anas platyrhynchos) using random access amplification and next generation sequencing technologies.Virol J. 2011 Oct 6;8:463. doi: 10.1186/1743-422X-8-463. Virol J. 2011. PMID: 21978491 Free PMC article.
-
Pathogenesis of two strains of avian paramyxovirus serotype 2, Yucaipa and Bangor, in chickens and turkeys.Avian Dis. 2010 Sep;54(3):1050-7. doi: 10.1637/9380-041910-Reg.1. Avian Dis. 2010. PMID: 20945787 Free PMC article.
References
-
- Alexander DJ. Newcastle disease and other avian Paramyxoviridae infections. In: Calnek BW, editor. Diseases of Poultry. Ames: Iowa State University Press; 1997. pp. 541–569.
-
- Alexander DJ. Avian Paramyxoviruses 2–9. In: Saif YM, editor. Diseases of poultry. 11th edn. Ames: Iowa State University Press; 2003. pp. 88–92.
-
- Alexander DJ, Collins MS. Pathogenecity of PMV-3/Parakeet/Netherland/449 /75 for chickens. Avian Pathol. 1982;11:179–185. - PubMed
-
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: Signal 3.0. J Mol Biol. 2004;340:783–795. - PubMed
-
- Biacchesi S, Skiadopoulos MH, Boivin G, Hanson CT, Murphy BR, Collins PL, Buchholz UJ. Genetic diversity between human metapneumovirus subgroups. Virology. 2003;315:1–9. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

