Structural and functional properties of the vesicular stomatitis virus nucleoprotein-RNA complex as revealed by proteolytic digestion
- PMID: 20206958
- PMCID: PMC2853252
- DOI: 10.1016/j.virol.2010.02.014
Structural and functional properties of the vesicular stomatitis virus nucleoprotein-RNA complex as revealed by proteolytic digestion
Abstract
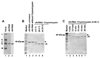
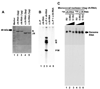
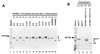
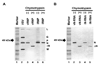
To gain insight into the structural and functional properties of the vesicular stomatitis virus nucleocapsid-RNA complex (vN-RNA), we analyzed it by treatment with proteolytic enzymes. Chymotrypsin treatment to the vN-RNA results in complete digestion of the C-terminal 86 amino acids of the N protein. The residual chymotrypsin resistant vN-RNA complex (vDeltaN-RNA) carrying N-terminal 336 amino acids of the N protein (DeltaN) was inactive in transcription. The DeltaN protein retained its capability to protect the genomic RNA from nuclease digestion but failed to interact to the P protein. Interestingly, addition of excess amount of P protein rendered the vN-RNA complex resistant to the chymotrypsin digestion. Finally, our data revealed that the recombinant N-RNA complex purified from bacteria (bN-RNA) is resistant to chymotrypsin digestion, suggesting that the C-terminal unstructured domain (C-loop) remains inaccessible to protease digestion. Detailed comparative analyses of the vN-RNA and vDeltaN-RNA are discussed.
Figures


Similar articles
-
Interaction of vesicular stomatitis virus P and N proteins: identification of two overlapping domains at the N terminus of P that are involved in N0-P complex formation and encapsidation of viral genome RNA.J Virol. 2007 Dec;81(24):13478-85. doi: 10.1128/JVI.01244-07. Epub 2007 Oct 3. J Virol. 2007. PMID: 17913815 Free PMC article.
-
The nucleocapsid of vesicular stomatitis virus.Sci China Life Sci. 2012 Apr;55(4):291-300. doi: 10.1007/s11427-012-4307-x. Epub 2012 May 9. Sci China Life Sci. 2012. PMID: 22566085 Review.
-
Solution structure of the C-terminal nucleoprotein-RNA binding domain of the vesicular stomatitis virus phosphoprotein.J Mol Biol. 2008 Oct 3;382(2):525-38. doi: 10.1016/j.jmb.2008.07.028. Epub 2008 Jul 16. J Mol Biol. 2008. PMID: 18657547
-
Structure of the vesicular stomatitis virus nucleoprotein-RNA complex.Science. 2006 Jul 21;313(5785):357-60. doi: 10.1126/science.1126953. Epub 2006 Jun 15. Science. 2006. PMID: 16778022
-
Structural disorder within the replicative complex of measles virus: functional implications.Virology. 2006 Jan 5;344(1):94-110. doi: 10.1016/j.virol.2005.09.025. Virology. 2006. PMID: 16364741 Review.
Cited by
-
Strategies for viral RNA stability: live long and prosper.Trends Genet. 2011 Jul;27(7):286-93. doi: 10.1016/j.tig.2011.04.003. Trends Genet. 2011. PMID: 21640425 Free PMC article. Review.
-
Complete study demonstrating the absence of rhabdovirus in a distinct Sf9 cell line.PLoS One. 2017 Apr 19;12(4):e0175633. doi: 10.1371/journal.pone.0175633. eCollection 2017. PLoS One. 2017. PMID: 28423032 Free PMC article.
References
-
- Albertini AA, Wernimont AK, Muziol T, Ravelli RB, Clapier CR, Schoehn G, Weissenhorn W, Ruigrok RW. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science. 2006;313(5785):360–363. - PubMed
-
- Banerjee AK, Barik S. Gene expression of vesicular stomatitis virus genome RNA. Virology. 1992;188(2):417–428. - PubMed
-
- Banerjee AK, Barik S, De BP. Gene expression of nonsegmented negative strand RNA viruses. Pharmacol Ther. 1991;51(1):47–70. - PubMed
-
- Banerjee AK, Roy J, Chattopadhyay DJ. Effect of the proteolytic digestion on the function of vesicular stomatitis virus ribonucleoprotein complex. J. Biosci. 1987b;11:515–523.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

