RNAi-dependent formation of heterochromatin and its diverse functions
- PMID: 20207534
- PMCID: PMC3005588
- DOI: 10.1016/j.gde.2010.02.003
RNAi-dependent formation of heterochromatin and its diverse functions
Abstract
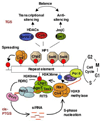
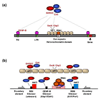
Expression profiling of eukaryotic genomes has revealed widespread transcription outside the confines of protein-coding genes, leading to production of antisense and non-coding RNAs (ncRNAs). Studies in Schizosaccharomyces pombe and multicellular organisms suggest that transcription and ncRNAs provide a framework for the assembly of heterochromatin, which has been linked to various chromosomal processes. In addition to gene regulation, heterochromatin is crucial for centromere function, cell fate determination as well as transcriptional and posttranscriptional silencing of repetitive DNA elements. Recently, heterochromatin factors have been shown to suppress antisense RNAs at euchromatic loci. These findings define conserved pathways that probably have major impact on the epigenetic regulation of eukaryotic genomes.
Published by Elsevier Ltd.
Figures
References
-
- Gingeras TR. Origin of phenotypes: genes and transcripts. Genome Res. 2007;17:682–690. - PubMed
-
- Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat. Struct. Mol. Biol. 2007;14:103–105. - PubMed
-
-
Dutrow N, Nix DA, Holt D, Milash B, Dalley B, Westbroek E, Parnell TJ, Cairns BR. Dynamic transcriptome of Schizosaccharomyces pombe shown by RNA-DNA hybrid mapping. Nat. Genet. 2008;40:977–986. References and show that major portions of the S. pombe genome including intergenic regions are transcribed. These studies also identify extensive antisense transcription, and induction of large numbers of non-coding RNAs under different growth and environmental conditions.
-
-
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

