Ring-shaped Rad51 paralog protein complexes bind Holliday junctions and replication forks as visualized by electron microscopy
- PMID: 20207730
- PMCID: PMC2859493
- DOI: 10.1074/jbc.M109.074286
Ring-shaped Rad51 paralog protein complexes bind Holliday junctions and replication forks as visualized by electron microscopy
Abstract
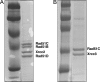
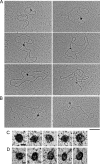
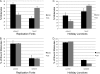
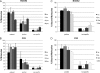
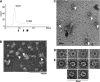
In mammals, there are five Rad51 paralogs that form two distinct complexes in vivo. One complex is composed of Rad51B-Rad51C-Rad51D-Xrcc2 (BCDX2) and the other Rad51C-Xrcc3 (CX3). We co-expressed and purified human BCDX2 and CX3 protein complexes from insect cells and investigated their binding preferences and structure using transmission electron microscopy (TEM). We visualized the binding of BCDX2 and CX3 to DNA templates containing replication forks and Holliday junctions, intermediates observed during DNA replication and recombination, respectively. We show that both complexes bind with exceptionally high specificity to the DNA junctions with little binding observed elsewhere on the DNAs. Further analysis of the structure of free or DNA-bound BCDX2 and CX3 complexes revealed a multimeric ring structure whose subunits are arranged into a flat disc around a central channel. This work provides the first EM visualization of BCDX2 and CX3 binding to Holliday junctions and forked DNAs and suggests the complexes form ring-shaped structures.
Figures

Similar articles
-
Structure and function of the RAD51B-RAD51C-RAD51D-XRCC2 tumour suppressor.Nature. 2023 Jul;619(7970):650-657. doi: 10.1038/s41586-023-06179-1. Epub 2023 Jun 21. Nature. 2023. PMID: 37344587 Free PMC article.
-
Mechanism of BCDX2-mediated RAD51 nucleation on short ssDNA stretches and fork DNA.Nucleic Acids Res. 2024 Oct 28;52(19):11738-11752. doi: 10.1093/nar/gkae770. Nucleic Acids Res. 2024. PMID: 39268578 Free PMC article.
-
Structural insights into BCDX2 complex function in homologous recombination.Nature. 2023 Jul;619(7970):640-649. doi: 10.1038/s41586-023-06219-w. Epub 2023 Jun 21. Nature. 2023. PMID: 37344589 Free PMC article.
-
Rad51 paralog complexes BCDX2 and CX3 act at different stages in the BRCA1-BRCA2-dependent homologous recombination pathway.Mol Cell Biol. 2013 Jan;33(2):387-95. doi: 10.1128/MCB.00465-12. Epub 2012 Nov 12. Mol Cell Biol. 2013. PMID: 23149936 Free PMC article.
-
RAD-ical New Insights into RAD51 Regulation.Genes (Basel). 2018 Dec 13;9(12):629. doi: 10.3390/genes9120629. Genes (Basel). 2018. PMID: 30551670 Free PMC article. Review.
Cited by
-
DNA-binding properties of T4 UvsY recombination mediator protein: polynucleotide wrapping promotes high-affinity binding to single-stranded DNA.Nucleic Acids Res. 2010 Aug;38(14):4821-33. doi: 10.1093/nar/gkq219. Epub 2010 Apr 5. Nucleic Acids Res. 2010. PMID: 20371513 Free PMC article.
-
DNA repair pathways in trypanosomatids: from DNA repair to drug resistance.Microbiol Mol Biol Rev. 2014 Mar;78(1):40-73. doi: 10.1128/MMBR.00045-13. Microbiol Mol Biol Rev. 2014. PMID: 24600040 Free PMC article. Review.
-
RAD51B Activity and Cell Cycle Regulation in Response to DNA Damage in Breast Cancer Cell Lines.Breast Cancer (Auckl). 2014 Oct 12;8:135-44. doi: 10.4137/BCBCR.S17766. eCollection 2014. Breast Cancer (Auckl). 2014. PMID: 25368520 Free PMC article.
-
The HsRAD51B-HsRAD51C stabilizes the HsRAD51 nucleoprotein filament.DNA Repair (Amst). 2013 Sep;12(9):723-32. doi: 10.1016/j.dnarep.2013.05.005. Epub 2013 Jun 28. DNA Repair (Amst). 2013. PMID: 23810717 Free PMC article.
-
Overexpression of Rad51C splice variants in colorectal tumors.Oncotarget. 2015 Apr 20;6(11):8777-87. doi: 10.18632/oncotarget.3209. Oncotarget. 2015. PMID: 25669972 Free PMC article.
References
-
- Jang Y. K., Jin Y. H., Shim Y. S., Kim M. J., Yoo E. J., Choi I. S., Lee J. S., Seong R. H., Hong S. H., Park S. D. (1996) Mol. Gen. Genet 251, 167–175 - PubMed
-
- Mazin A. V., Bornarth C. J., Solinger J. A., Heyer W. D., Kowalczykowski S. C. (2000) Mol. Cell 6, 583–592 - PubMed
-
- Benson F. E., Baumann P., West S. C. (1998) Nature 391, 401–404 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

