Mechanistic insight into the function of the C-terminal PKD domain of the collagenolytic serine protease deseasin MCP-01 from deep sea Pseudoalteromonas sp. SM9913: binding of the PKD domain to collagen results in collagen swelling but does not unwind the collagen triple helix
- PMID: 20207733
- PMCID: PMC2863174
- DOI: 10.1074/jbc.M109.087023
Mechanistic insight into the function of the C-terminal PKD domain of the collagenolytic serine protease deseasin MCP-01 from deep sea Pseudoalteromonas sp. SM9913: binding of the PKD domain to collagen results in collagen swelling but does not unwind the collagen triple helix
Abstract
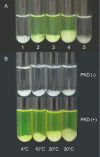
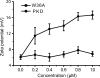
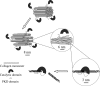
Deseasin MCP-01 is a bacterial collagenolytic serine protease. Its catalytic domain alone can degrade collagen, and its C-terminal PKD domain is a collagen-binding domain (CBD) that can improve the collagenolytic efficiency of the catalytic domain by an unknown mechanism. Here, scanning electron microscopy (SEM), atomic force microscopy (AFM), zeta potential, and circular dichroism spectroscopy were used to clarify the functional mechanism of the PKD domain in MCP-01 collagenolysis. The PKD domain observably swelled insoluble collagen. Its collagen-swelling ability and its improvement to the collagenolysis of the catalytic domain are both temperature-dependent. SEM observation showed the PKD domain swelled collagen fascicles with an increase of their diameter from 5.3 mum to 8.8 mum after 1 h of treatment, and the fibrils forming the fascicles were dispersed. AFM observation directly showed that the PKD domain bound collagen, swelled the microfibrils, and exposed the monomers. The PKD mutant W36A neither bound collagen nor disturbed its structure. Zeta potential results demonstrated that PKD treatment increased the net positive charges of the collagen surface. PKD treatment caused no change in the content or the thermostability of the collagen triple helix. Furthermore, the PKD-treated collagen could not be degraded by gelatinase. Therefore, though the triple helix monomers were exposed, the PKD domain could not unwind the collagen triple helix. Our study reveals the functional mechanism of the PKD domain of the collagenolytic serine protease MCP-01 in collagen degradation, which is distinct from that of the CBDs of mammalian matrix metalloproteases.
Figures






References
-
- Gluecksmann-Kuis M. A., Tayber O., Woolf E. A., Bougueleret L., Deng N., Alperin G. D., Iris F., Hawkins F. (1995) Cell 81, 289–298
-
- Jing H., Takagi J., Liu J. H., Lindgren S., Zhang R. G., Joachimiak A., Wang J. H., Springer T. A. (2002) Structure 10, 1453–1464 - PubMed
-
- Perrakis A., Tews I., Dauter Z., Oppenheim A. B., Chet I., Wilson K. S., Vorgias C. E. (1994) Structure 2, 1169–1180 - PubMed
-
- Orikoshi H., Nakayama S., Hanato C., Miyamoto K., Tsujibo H. (2005) J. Appl. Microbiol. 99, 551–557 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

