Ternary complex of transforming growth factor-beta1 reveals isoform-specific ligand recognition and receptor recruitment in the superfamily
- PMID: 20207738
- PMCID: PMC2863181
- DOI: 10.1074/jbc.M109.079921
Ternary complex of transforming growth factor-beta1 reveals isoform-specific ligand recognition and receptor recruitment in the superfamily
Abstract
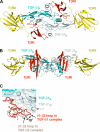
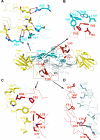
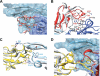
Transforming growth factor (TGF)-beta1, -beta2, and -beta3 are 25-kDa homodimeric polypeptides that play crucial nonoverlapping roles in embryogenesis, tissue development, carcinogenesis, and immune regulation. Here we report the 3.0-A resolution crystal structure of the ternary complex between human TGF-beta1 and the extracellular domains of its type I and type II receptors, TbetaRI and TbetaRII. The TGF-beta1 ternary complex structure is similar to previously reported TGF-beta3 complex except with a 10 degrees rotation in TbetaRI docking orientation. Quantitative binding studies showed distinct kinetics between the receptors and the isoforms of TGF-beta. TbetaRI showed significant binding to TGF-beta2 and TGF-beta3 but not TGF-beta1, and the binding to all three isoforms of TGF-beta was enhanced considerably in the presence of TbetaRII. The preference of TGF-beta2 to TbetaRI suggests a variation in its receptor recruitment in vivo. Although TGF-beta1 and TGF-beta3 bind and assemble their ternary complexes in a similar manner, their structural differences together with differences in the affinities and kinetics of their receptor binding may underlie their unique biological activities. Structural comparisons revealed that the receptor-ligand pairing in the TGF-beta superfamily is dictated by unique insertions, deletions, and disulfide bonds rather than amino acid conservation at the interface. The binding mode of TbetaRII on TGF-beta is unique to TGF-betas, whereas that of type II receptor for bone morphogenetic protein on bone morphogenetic protein appears common to all other cytokines in the superfamily. Further, extensive hydrogen bonds and salt bridges are present at the high affinity cytokine-receptor interfaces, whereas hydrophobic interactions dominate the low affinity receptor-ligand interfaces.
Figures



Similar articles
-
TbetaR-II discriminates the high- and low-affinity TGF-beta isoforms via two hydrogen-bonded ion pairs.Biochemistry. 2009 Mar 17;48(10):2146-55. doi: 10.1021/bi8019004. Biochemistry. 2009. PMID: 19161338 Free PMC article.
-
Transforming growth factor-beta (TGF-beta) binding to the extracellular domain of the type II TGF-beta receptor: receptor capture on a biosensor surface using a new coiled-coil capture system demonstrates that avidity contributes significantly to high affinity binding.J Mol Biol. 2003 May 16;328(5):1173-83. doi: 10.1016/s0022-2836(03)00360-7. J Mol Biol. 2003. PMID: 12729750
-
In the absence of type III receptor, the transforming growth factor (TGF)-beta type II-B receptor requires the type I receptor to bind TGF-beta2.J Biol Chem. 2004 May 21;279(21):22765-72. doi: 10.1074/jbc.M401350200. Epub 2004 Mar 2. J Biol Chem. 2004. PMID: 14996829
-
Role of TGF-β3 in the regulation of immune responses.Clin Exp Rheumatol. 2015 Jul-Aug;33(4 Suppl 92):S63-9. Epub 2015 Oct 12. Clin Exp Rheumatol. 2015. PMID: 26457612 Review.
-
Analysing the relevance of TGF-β and its regulators in cervical cancer to identify therapeutic and diagnostic markers.Gene. 2025 Feb 20;938:149166. doi: 10.1016/j.gene.2024.149166. Epub 2024 Dec 18. Gene. 2025. PMID: 39701195 Review.
Cited by
-
Oleuropein Protects Against Cerebral Ischemia Injury in Rats: Molecular Docking, Biochemical and Histological Findings.Neurochem Res. 2021 Aug;46(8):2131-2142. doi: 10.1007/s11064-021-03351-9. Epub 2021 May 18. Neurochem Res. 2021. PMID: 34008118
-
Members of the DAN family are BMP antagonists that form highly stable noncovalent dimers.J Mol Biol. 2012 Dec 14;424(5):313-27. doi: 10.1016/j.jmb.2012.10.003. Epub 2012 Oct 9. J Mol Biol. 2012. PMID: 23063586 Free PMC article.
-
Structural Adaptation in Its Orphan Domain Engenders Betaglycan with an Alternate Mode of Growth Factor Binding Relative to Endoglin.Structure. 2019 Sep 3;27(9):1427-1442.e4. doi: 10.1016/j.str.2019.06.010. Epub 2019 Jul 18. Structure. 2019. PMID: 31327662 Free PMC article.
-
Network Pharmacology Research Indicates that Wu-Mei-Wan Treats Obesity by Inhibiting Th17 Cell Differentiation and Alleviating Metabolic Inflammation.Comb Chem High Throughput Screen. 2023;26(1):30-48. doi: 10.2174/1386207325666220221121919. Comb Chem High Throughput Screen. 2023. PMID: 35189797
-
New strategy for high-level expression and purification of biologically active monomeric TGF-β1/C77S in Escherichia coli.Mol Biotechnol. 2015 Feb;57(2):160-71. doi: 10.1007/s12033-014-9812-7. Mol Biotechnol. 2015. PMID: 25370824
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

