bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis
- PMID: 20207753
- PMCID: PMC2861475
- DOI: 10.1105/tpc.109.072173
bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis
Abstract
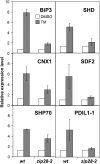
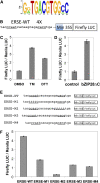
Stress agents known to elicit the unfolded protein response in Arabidopsis thaliana upregulate the expression of a constellation of genes dependent on the membrane-associated basic domain/leucine zipper (bZIP) transcription factor, bZIP28. Among the stress-activated genes, a consensus promoter sequence corresponding to the endoplasmic reticulum (ER) stress-responsive element I (ERSE-I), CCAAT-N10-CACG, was identified. Disruption of either the CCAAT or CACG subelement in ERSE-I resulted in reduction of the transcriptional response to ER stress. bZIP28 forms homo- and heterodimers with other bZIP TF family members (in subgroup D) and interacts with CCAAT box binding factors, heterotrimeric factors composed of NF-Y subunits. Arabidopsis encodes 36 NF-Y subunits, and it was found that subunits NF-YB3 and -YC2 interact with bZIP28 and NF-YA4, respectively, in a yeast three-hybrid system. A transcriptional complex containing bZIP28 and the above-mentioned three NF-Y subunits was assembled in vitro on DNA containing ERSE-I. bZIP28, on its own, binds to the CACG subelement in ERSE-I to form a smaller complex I, and in combination with the NF-Y subunits above, bZIP28 assembles into a larger transcriptional complex (complex II). bZIP28 was shown to interact with NF-Y subunits in vivo in bimolecular fluorescence complementation analyses and in coimmunoprecipitation assays. Treatment of seedlings with ER stress agents led to the upregulation of NF-YC2 and the relocation of NF-YB3 from the cytoplasm to the nucleus. Thus, in response to ER stress, bZIP28 is mobilized by proteolysis and recruits NF-Y subunits to form a transcriptional complex that upregulates the expression of ER stress-induced genes.
Figures








References
-
- Ben-Naim O., Eshed R., Parnis A., Teper-Bamnolker P., Shalit A., Coupland G., Samach A., Lifschitz E. (2006). The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J. 46: 462–476 - PubMed
-
- Bernales S., Papa F.R., Walter P. (2006). Intracellular signaling by the unfolded protein response. Annu. Rev. Cell Dev. Biol. 22: 487–508 - PubMed
-
- Chen X., Shen J., Prywes R. (2002). The luminal domain of ATF6 senses endoplasmic reticulum (ER) stress and causes translocation of ATF6 from the ER to the Golgi. J. Biol. Chem. 277: 13045–13052 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

