Mitochondrial DNA heteroplasmy in Candida glabrata after mitochondrial transformation
- PMID: 20207853
- PMCID: PMC2863964
- DOI: 10.1128/EC.00349-09
Mitochondrial DNA heteroplasmy in Candida glabrata after mitochondrial transformation
Abstract
Genetic manipulation of mitochondrial DNA (mtDNA) is the most direct method for investigating mtDNA, but until now, this has been achieved only in the diploid yeast Saccharomyces cerevisiae. In this study, the ATP6 gene on mtDNA of the haploid yeast Candida glabrata (Torulopsis glabrata) was deleted by biolistic transformation of DNA fragments with a recoded ARG8(m) mitochondrial genetic marker, flanked by homologous arms to the ATP6 gene. Transformants were identified by arginine prototrophy. However, in the transformants, the original mtDNA was not lost spontaneously, even under arginine selective pressure. Moreover, the mtDNA transformants selectively lost the transformed mtDNA under aerobic conditions. The mtDNA heteroplasmy in the transformants was characterized by PCR, quantitative PCR, and Southern blotting, showing that the heteroplasmy was relatively stable in the absence of arginine. Aerobic conditions facilitated the loss of the original mtDNA, and anaerobic conditions favored loss of the transformed mtDNA. Moreover, detailed investigations showed that increases in reactive oxygen species in mitochondria lacking ATP6, along with their equal cell division, played important roles in determining the dynamics of heteroplasmy. Based on our analysis of mtDNA heteroplasmy in C. glabrata, we were able to generate homoplasmic Deltaatp6 mtDNA strains.
Figures
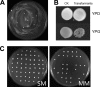

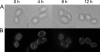
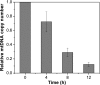
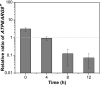

Similar articles
-
Method to purify mitochondrial DNA directly from yeast total DNA.Plasmid. 2010 Nov;64(3):196-9. doi: 10.1016/j.plasmid.2010.06.005. Epub 2010 Jun 29. Plasmid. 2010. PMID: 20600282
-
Mitochondrial fusion increases the mitochondrial DNA copy number in budding yeast.Genes Cells. 2011 May;16(5):527-44. doi: 10.1111/j.1365-2443.2011.01504.x. Epub 2011 Apr 5. Genes Cells. 2011. PMID: 21463454
-
Inactivation of the 20S proteasome maturase, Ump1p, leads to the instability of mtDNA in Saccharomyces cerevisiae.Mutat Res. 2009 Oct 2;669(1-2):95-103. doi: 10.1016/j.mrfmmm.2009.05.008. Epub 2009 May 23. Mutat Res. 2009. PMID: 19467248
-
Biolistic transformation of the yeast Saccharomyces cerevisiae mitochondrial DNA.IUBMB Life. 2023 Dec;75(12):972-982. doi: 10.1002/iub.2769. Epub 2023 Jul 20. IUBMB Life. 2023. PMID: 37470229 Review.
-
Running on empty: does mitochondrial DNA mutation limit replicative lifespan in yeast?: Mutations that increase the division rate of cells lacking mitochondrial DNA also extend replicative lifespan in Saccharomyces cerevisiae.Bioessays. 2011 Oct;33(10):742-8. doi: 10.1002/bies.201100050. Epub 2011 Aug 9. Bioessays. 2011. PMID: 21826691 Review.
Cited by
-
Mechanisms of multidrug resistance caused by an Ipi1 mutation in the fungal pathogen Candida glabrata.Nat Commun. 2025 Jan 25;16(1):1023. doi: 10.1038/s41467-025-56269-z. Nat Commun. 2025. PMID: 39863615 Free PMC article.
-
Biolistic Transformation of Chlamydomonas reinhardtii and Saccharomyces cerevisiae Mitochondria.Methods Mol Biol. 2023;2615:345-364. doi: 10.1007/978-1-0716-2922-2_24. Methods Mol Biol. 2023. PMID: 36807803
-
Evidence for Persistent Heteroplasmy and Ancient Recombination in the Mitochondrial Genomes of the Edible Yellow Chanterelles From Southwestern China and Europe.Front Microbiol. 2021 Jul 14;12:699598. doi: 10.3389/fmicb.2021.699598. eCollection 2021. Front Microbiol. 2021. PMID: 34335532 Free PMC article.
-
Transfection of plant mitochondria and in organello gene integration.Nucleic Acids Res. 2011 Sep 1;39(17):e115. doi: 10.1093/nar/gkr517. Epub 2011 Jun 28. Nucleic Acids Res. 2011. PMID: 21715377 Free PMC article.
-
DNA delivery to mitochondria: sequence specificity and energy enhancement.Pharm Res. 2011 Nov;28(11):2871-82. doi: 10.1007/s11095-011-0516-4. Epub 2011 Jul 12. Pharm Res. 2011. PMID: 21748538
References
-
- Adrio J. L., Demain A. L. 2006. Genetic improvement of processes yielding microbial products. FEMS Microbiol. Rev. 30:187–214 - PubMed
-
- Berger K. H., Yaffe M. P. 2000. Mitochondrial DNA inheritance in Saccharomyces cerevisiae. Trends Microbiol. 8:508–513 - PubMed
-
- Bialkova A., Subik J. 2006. Biology of the pathogenic yeast Candida glabrata. Folia Microbiol. (Praha) 51:3–20 - PubMed
-
- Bonnefoy N., Remacle C., Fox T. D. 2007. Genetic transformation of Saccharomyces cerevisiae and Chlamydomonas reinhardtii mitochondria. Methods Cell Biol. 80:525–548 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

