Kinetic cooperativity in Escherichia coli 30S ribosomal subunit reconstitution reveals additional complexity in the assembly landscape
- PMID: 20207951
- PMCID: PMC2851750
- DOI: 10.1073/pnas.0912007107
Kinetic cooperativity in Escherichia coli 30S ribosomal subunit reconstitution reveals additional complexity in the assembly landscape
Abstract
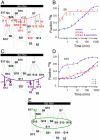
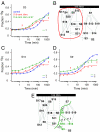
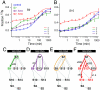
The Escherichia coli 30S ribosomal subunit self-assembles in vitro in a hierarchical manner, with the RNA binding by proteins enabled by the prior binding of others under equilibrium conditions. Early 16S rRNA binding proteins also bind faster than late-binding proteins, but the specific causes for the slow binding of late proteins remain unclear. Previously, a pulse-chase monitored by quantitative mass spectrometry method was developed for monitoring 30S subunit assembly kinetics, and here a modified experimental scheme was used to probe kinetic cooperativity by including a step where subsets of ribosomal proteins bind and initiate assembly prior to the pulse-chase kinetics. In this work, 30S ribosomal subunit kinetic reconstitution experiments revealed that thermodynamic dependency does not always correlate with kinetic cooperativity. Some folding transitions that cause subsequent protein binding to be more energetically favorable do not result in faster protein binding. Although 3(') domain primary protein S7 is required for RNA binding by both proteins S9 and S19, prior binding of S7 accelerates the binding of S9, but not S19, indicating there is an additional mechanistic step required for S19 to bind. Such data on kinetic cooperativity and the presence of multiphasic assembly kinetics reveal complexity in the assembly landscape that was previously hidden.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
The effect of ribosome assembly cofactors on in vitro 30S subunit reconstitution.J Mol Biol. 2010 Apr 23;398(1):1-7. doi: 10.1016/j.jmb.2010.02.036. Epub 2010 Feb 24. J Mol Biol. 2010. PMID: 20188109 Free PMC article.
-
Assembly of the 30S ribosomal subunit.Q Rev Biophys. 2005 Nov;38(4):397-403. doi: 10.1017/S0033583506004264. Q Rev Biophys. 2005. PMID: 16934171
-
Assembly of the 30S ribosomal subunit: positioning ribosomal protein S13 in the S7 assembly branch.RNA. 2004 Dec;10(12):1861-6. doi: 10.1261/rna.7130504. Epub 2004 Nov 3. RNA. 2004. PMID: 15525707 Free PMC article.
-
A complex assembly landscape for the 30S ribosomal subunit.Annu Rev Biophys. 2009;38:197-215. doi: 10.1146/annurev.biophys.050708.133615. Annu Rev Biophys. 2009. PMID: 19416066 Free PMC article. Review.
-
Structural insights into cell cycle control by essential GTPase Era.Postepy Biochem. 2016;62(3):335-342. Postepy Biochem. 2016. PMID: 28132488 Free PMC article. Review.
Cited by
-
Structural insights into the assembly of the 30S ribosomal subunit in vivo: functional role of S5 and location of the 17S rRNA precursor sequence.Protein Cell. 2014 May;5(5):394-407. doi: 10.1007/s13238-014-0044-1. Epub 2014 Mar 28. Protein Cell. 2014. PMID: 24671761 Free PMC article.
-
RNA folding pathways and the self-assembly of ribosomes.Acc Chem Res. 2011 Dec 20;44(12):1312-9. doi: 10.1021/ar2000474. Epub 2011 Jun 29. Acc Chem Res. 2011. PMID: 21714483 Free PMC article.
-
Assembly constraints drive co-evolution among ribosomal constituents.Nucleic Acids Res. 2015 Jun 23;43(11):5352-63. doi: 10.1093/nar/gkv448. Epub 2015 May 8. Nucleic Acids Res. 2015. PMID: 25956649 Free PMC article.
-
Evolution of protein-coupled RNA dynamics during hierarchical assembly of ribosomal complexes.Nat Commun. 2017 Sep 8;8(1):492. doi: 10.1038/s41467-017-00536-1. Nat Commun. 2017. PMID: 28887451 Free PMC article.
-
Transcription Increases the Cooperativity of Ribonucleoprotein Assembly.Cell. 2019 Nov 27;179(6):1370-1381.e12. doi: 10.1016/j.cell.2019.11.007. Epub 2019 Nov 21. Cell. 2019. PMID: 31761536 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

